Single-cell RNA sequencing profiling of mouse endothelial cells in response to pulmonary arterial hypertension
- PMID: 34528097
- PMCID: PMC9400412
- DOI: 10.1093/cvr/cvab296
Single-cell RNA sequencing profiling of mouse endothelial cells in response to pulmonary arterial hypertension
Abstract
Aims: Endothelial cell (EC) dysfunction drives the initiation and pathogenesis of pulmonary arterial hypertension (PAH). We aimed to characterize EC dynamics in PAH at single-cell resolution.
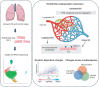
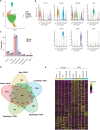
Methods and results: We carried out single-cell RNA sequencing (scRNA-seq) of lung ECs isolated from an EC lineage-tracing mouse model in Control and SU5416/hypoxia-induced PAH conditions. EC populations corresponding to distinct lung vessel types, including two discrete capillary populations, were identified in both Control and PAH mice. Differential gene expression analysis revealed global PAH-induced EC changes that were confirmed by bulk RNA-seq. This included upregulation of the major histocompatibility complex class II pathway, supporting a role for ECs in the inflammatory response in PAH. We also identified a PAH response specific to the second capillary EC population including upregulation of genes involved in cell death, cell motility, and angiogenesis. Interestingly, four genes with genetic variants associated with PAH were dysregulated in mouse ECs in PAH. To compare relevance across PAH models and species, we performed a detailed analysis of EC heterogeneity and response to PAH in rats and humans through whole-lung PAH scRNA-seq datasets, revealing that 51% of up-regulated mouse genes were also up-regulated in rat or human PAH. We identified promising new candidates to target endothelial dysfunction including CD74, the knockdown of which regulates EC proliferation and barrier integrity in vitro. Finally, with an in silico cell ordering approach, we identified zonation-dependent changes across the arteriovenous axis in mouse PAH and showed upregulation of the Serine/threonine-protein kinase Sgk1 at the junction between the macro- and microvasculature.
Conclusion: This study uncovers PAH-induced EC transcriptomic changes at a high resolution, revealing novel targets for potential therapeutic candidate development.
Keywords: Endothelial cells; PAH; Pulmonary hypertension; Single-cell RNA-seq.
© The Author(s) 2021. Published by Oxford University Press on behalf of the European Society of Cardiology.
Conflict of interest statement
Conflict of interest: S.Y.C. has served as a consultant for United Therapeutics, has held research grants from Actelion and Pfizer, and filed patent applications regarding drug development in pulmonary hypertension. S.Y.C. is a director, officer, and shareholder of Synhale Therapeutics. The other authors declare no competing interests. This manuscript was handled by Consulting Editor Henning Morawietz.
Figures






Comment in
-
Response to: Are endothelial cell proliferation and mesenchymal transition as distinguishing characteristics of 3-week Sugen5416/hypoxia mice model?Cardiovasc Res. 2023 Jul 6;119(8):e142-e143. doi: 10.1093/cvr/cvad075. Cardiovasc Res. 2023. PMID: 37170759 Free PMC article. No abstract available.
-
Are endothelial cell proliferation and mesenchymal transition as distinguishing characteristics of 3-week Sugen5416/hypoxia mice model?Cardiovasc Res. 2023 Jul 6;119(8):e140-e141. doi: 10.1093/cvr/cvad074. Cardiovasc Res. 2023. PMID: 37183500 No abstract available.
References
-
- Lau EMT, Giannoulatou E, Celermajer DS, Humbert M.. Epidemiology and treatment of pulmonary arterial hypertension. Nat Rev Cardiol 2017;14:603–614. - PubMed
-
- Peacock AJ, Murphy NF, McMurray JJ, Caballero L, Stewart S.. An epidemiological study of pulmonary arterial hypertension. Eur Respir J 2007;30:104–109. - PubMed
-
- Ciuclan L, Bonneau O, Hussey M, Duggan N, Holmes AM, Good R, Stringer R, Jones P, Morrell NW, Jarai G, Walker C, Westwick J, Thomas M.. A novel murine model of severe pulmonary arterial hypertension. Am J Respir Crit Care Med 2011;184:1171–1182. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

