Epithelial memory of inflammation limits tissue damage while promoting pancreatic tumorigenesis
- PMID: 34529467
- PMCID: PMC9733946
- DOI: 10.1126/science.abj0486
Epithelial memory of inflammation limits tissue damage while promoting pancreatic tumorigenesis
Abstract
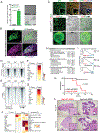
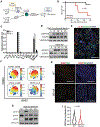
Inflammation is a major risk factor for pancreatic ductal adenocarcinoma (PDAC). When occurring in the context of pancreatitis, KRAS mutations accelerate tumor development in mouse models. We report that long after its complete resolution, a transient inflammatory event primes pancreatic epithelial cells to subsequent transformation by oncogenic KRAS. Upon recovery from acute inflammation, pancreatic epithelial cells display an enduring adaptive response associated with sustained transcriptional and epigenetic reprogramming. Such adaptation enables the reactivation of acinar-to-ductal metaplasia (ADM) upon subsequent inflammatory events, thereby limiting tissue damage through a rapid decrease of zymogen production. We propose that because activating mutations of KRAS maintain an irreversible ADM, they may be beneficial and under strong positive selection in the context of recurrent pancreatitis.
Conflict of interest statement
Competing interests
A.V., A.C., E.D.P and I.H. are inventors on a patent pertaining to the use of ADM inducers to treat pancreatitis and prevent pancreatic cancer.
Figures




Comment in
-
Under pressure.Nat Rev Cancer. 2021 Dec;21(12):743. doi: 10.1038/s41568-021-00416-3. Nat Rev Cancer. 2021. PMID: 34611351 No abstract available.
References
-
- Mantovani A, Allavena P, Sica A, Balkwill F, Cancer-related inflammation. Nature 454, 436–444 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

