A TP53 mutation model for the prediction of prognosis and therapeutic responses in head and neck squamous cell carcinoma
- PMID: 34530752
- PMCID: PMC8447564
- DOI: 10.1186/s12885-021-08765-w
A TP53 mutation model for the prediction of prognosis and therapeutic responses in head and neck squamous cell carcinoma
Abstract
Background: Tumor protein p53 (TP53) is the most frequently mutated gene in head and neck squamous cell carcinoma (HNSC), and TP53 mutations are associated with inhibited immune signatures and poor prognosis. We established a TP53 mutation associated risk score model to evaluate the prognosis and therapeutic responses of patients with HNSC.
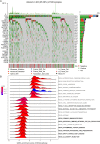
Methods: Differentially expressed genes between patients with and without TP53 mutations were determined by using data from the HNSC cohort in The Cancer Genome Atlas database. Patients with HNSC were divided into high- and low-risk groups based on a prognostic risk score that was generated from ten TP53 mutation associated genes via the multivariate Cox regression model.
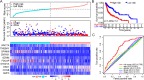
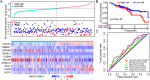
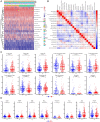
Results: TP53 was the most common mutant gene in HNSC, and TP53 mutations were associated with immunogenic signatures, including the infiltration of immune cells and expression of immune-associated genes. Patients in the high-risk group had significantly poorer overall survival than those in the low-risk group. The high-risk group showed less response to anti-programmed cell death protein 1 (PD-1) therapy but high sensitivity to some chemotherapies.
Conclusion: The risk score based on our TP53 mutation model was associated with poorer survival and could act as a specific predictor for assessing prognosis and therapeutic response in patients with HNSC.
Keywords: Chemotherapy; Head and neck squamous cell carcinoma; Immunotherapy; Prognosis; TP53 mutation; Therapeutic response.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures


References
-
- Global Burden of Disease Cancer C. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3(4):524–548. doi: 10.1001/jamaoncol.2016.5688. - DOI - PMC - PubMed
-
- Miyauchi S, Kim SS, Pang J, Gold KA, Gutkind JS, Califano JA, Mell LK, Cohen EEW, Sharabi AB. Immune modulation of head and neck squamous cell carcinoma and the tumor microenvironment by conventional therapeutics. Clin Cancer Res. 2019;25(14):4211–4223. doi: 10.1158/1078-0432.CCR-18-0871. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

