The biogeographic differentiation of algal microbiomes in the upper ocean from pole to pole
- PMID: 34531387
- PMCID: PMC8446083
- DOI: 10.1038/s41467-021-25646-9
The biogeographic differentiation of algal microbiomes in the upper ocean from pole to pole
Abstract
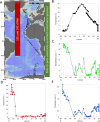
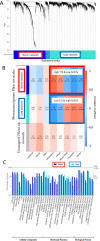
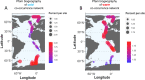
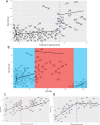
Eukaryotic phytoplankton are responsible for at least 20% of annual global carbon fixation. Their diversity and activity are shaped by interactions with prokaryotes as part of complex microbiomes. Although differences in their local species diversity have been estimated, we still have a limited understanding of environmental conditions responsible for compositional differences between local species communities on a large scale from pole to pole. Here, we show, based on pole-to-pole phytoplankton metatranscriptomes and microbial rDNA sequencing, that environmental differences between polar and non-polar upper oceans most strongly impact the large-scale spatial pattern of biodiversity and gene activity in algal microbiomes. The geographic differentiation of co-occurring microbes in algal microbiomes can be well explained by the latitudinal temperature gradient and associated break points in their beta diversity, with an average breakpoint at 14 °C ± 4.3, separating cold and warm upper oceans. As global warming impacts upper ocean temperatures, we project that break points of beta diversity move markedly pole-wards. Hence, abrupt regime shifts in algal microbiomes could be caused by anthropogenic climate change.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

