A forward genetic screen identifies modifiers of rocaglate responsiveness
- PMID: 34531456
- PMCID: PMC8445955
- DOI: 10.1038/s41598-021-97765-8
A forward genetic screen identifies modifiers of rocaglate responsiveness
Abstract
Rocaglates are a class of eukaryotic translation initiation inhibitors that are being explored as chemotherapeutic agents. They function by targeting eukaryotic initiation factor (eIF) 4A, an RNA helicase critical for recruitment of the 40S ribosome (and associated factors) to mRNA templates. Rocaglates perturb eIF4A activity by imparting a gain-of-function activity to eIF4A and mediating clamping to RNA. To appreciate how rocaglates could best be enabled in the clinic, an understanding of resistance mechanisms is important, as this could inform on strategies to bypass such events as well as identify responsive tumor types. Here, we report on the results of a positive selection, ORFeome screen aimed at identifying cDNAs capable of conferring resistance to rocaglates. Two of the most potent modifiers of rocaglate response identified were the transcription factors FOXP3 and NR1I3, both of which have been implicated in ABCB1 regulation-the gene encoding P-glycoprotein (Pgp). Pgp has previously been implicated in conferring resistance to silvestrol, a naturally occurring rocaglate, and we show here that this extends to additional synthetic rocaglate derivatives. In addition, FOXP3 and NR1I3 impart a multi-drug resistant phenotype that is reversed upon inhibition of Pgp, suggesting a potential therapeutic combination strategy.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

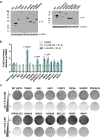

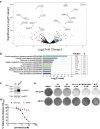

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

