Using host genetics to infer the global spread and evolutionary history of HCV subtype 3a
- PMID: 34532064
- PMCID: PMC8438900
- DOI: 10.1093/ve/veab065
Using host genetics to infer the global spread and evolutionary history of HCV subtype 3a
Abstract
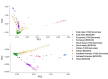
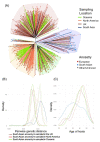
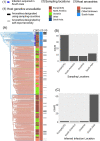
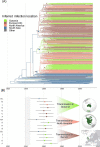
Studies have shown that hepatitis C virus subtype 3a (HCV-3a) is likely to have been circulating in South Asia before its global spread. However, the time and route of this dissemination remain unclear. For the first time, we generated host and virus genome-wide data for more than 500 patients infected with HCV-3a from the UK, North America, Australia, and New Zealand. We used the host genomic data to infer the ancestry of the patients and used this information to investigate the epidemic history of HCV-3a. We observed that viruses from hosts of South Asian ancestry clustered together near the root of the tree, irrespective of the sampling country, and that they were more diverse than viruses from other host ancestries. We hypothesized that South Asian hosts are more likely to have been infected in South Asia and used the inferred host ancestries to distinguish between the location where the infection was acquired and where the sample was taken. Next, we inferred that three independent transmission events resulted in the spread of the virus from South Asia to the UK, North America, and Oceania. This initial spread happened during or soon after the end of World War II. This was subsequently followed by many independent transmissions between the UK, North America, and Oceania. Using both host and virus genomic information can be highly informative in studying the virus epidemic history, especially in the context of chronic infections where migration histories need to be accounted for.
Keywords: HCV; evolution; host–virus genetics; phylogenetics; phylogeography.
© The Author(s) 2021. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
References
-
- British Government Act (1948), <https://www.legislation.gov.uk/ukpga/Geo6/11-12/56/enacted> accessed 5 Jul 2021.
Grants and funding
LinkOut - more resources
Full Text Sources

