Detection and evolution of SARS-CoV-2 coronavirus variants of concern with mass spectrometry
- PMID: 34532764
- PMCID: PMC8445501
- DOI: 10.1007/s00216-021-03649-1
Detection and evolution of SARS-CoV-2 coronavirus variants of concern with mass spectrometry
Abstract
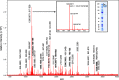
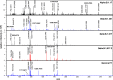
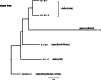
Mass mapping using high-resolution mass spectrometry has been applied to identify and rapidly distinguish SARS-CoV-2 coronavirus strains across five major variants of concern. Deletions or mutations within the surface spike protein across these variants, which originated in the UK, South Africa, Brazil and India (known as the alpha, beta, gamma and delta variants respectively), lead to associated mass differences in the mass maps. Peptides of unique mass have thus been determined that can be used to identify and distinguish the variants. The same mass map profiles are also utilized to construct phylogenetic trees, without the need for protein (or gene) sequences or their alignment, in order to chart and study viral evolution. The combined strategy offers advantages over conventional PCR-based gene-based approaches exploiting the ease with which protein mass maps can be generated and the speed and sensitivity of mass spectrometric analysis.
Keywords: Coronavirus; Evolution; Mass spectrometry; SARS-CoV-2; Variants; Virus.
© 2021. Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

