The Detrimental Role of Regulatory T Cells in Nonalcoholic Steatohepatitis
- PMID: 34532981
- PMCID: PMC8793993
- DOI: 10.1002/hep4.1807
The Detrimental Role of Regulatory T Cells in Nonalcoholic Steatohepatitis
Abstract
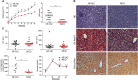
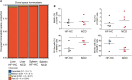
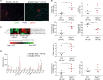
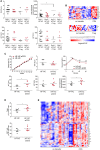
Nonalcoholic steatohepatitis (NASH) is induced by steatosis and metabolic inflammation. While involvement of the innate immune response has been shown, the role of the adaptive immune response in NASH remains controversial. Likewise, the role of regulatory T cells (Treg) in NASH remains unclear although initial clinical trials aim to target these regulatory responses. High-fat high-carbohydrate (HF-HC) diet feeding of NASH-resistant BALB/c mice as well as the corresponding recombination activating 1 (Rag)-deficient strain was used to induce NASH and to study the role of the adaptive immune response. HF-HC diet feeding induced strong activation of intrahepatic T cells in BALB/c mice, suggesting an antigen-driven effect. In contrast, the effects of the absence of the adaptive immune response was notable. NASH in BALB/c Rag1-/- mice was substantially worsened and accompanied by a sharp increase of M1-like macrophage numbers. Furthermore, we found an increase in intrahepatic Treg numbers in NASH, but either adoptive Treg transfer or anti-cluster of differentiation (CD)3 therapy unexpectedly increased steatosis and the alanine aminotransferase level without otherwise affecting NASH. Conclusion: Although intrahepatic T cells were activated and marginally clonally expanded in NASH, these effects were counterbalanced by increased Treg numbers. The ablation of adaptive immunity in murine NASH led to marked aggravation of NASH, suggesting that Tregs are not regulators of metabolic inflammation but rather enhance it.
© 2021 The Authors. Hepatology Communications published by Wiley Periodicals LLC on behalf of American Association for the Study of Liver Diseases.
Figures



References
-
- Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease‐meta‐analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73‐84. - PubMed
-
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11‐20. - PubMed
-
- Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol 2013;10:330‐344. - PubMed
-
- Wong RJ, Aguilar M, Cheung R, Perumpail RB, Harrison SA, Younossi ZM, et al. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology 2015;148:547‐555. - PubMed
-
- Golabi P, Bush H, Stepanova M, Locklear CT, Jacobson IM, Mishra A, et al. Liver transplantation (LT) for cryptogenic cirrhosis (CC) and nonalcoholic steatohepatitis (NASH) cirrhosis: data from the Scientific Registry of Transplant Recipients (SRTR): 1994 to 2016. Medicine (Baltimore) 2018;97:e11518. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

