Critical assessment of methods of protein structure prediction (CASP)-Round XIV
- PMID: 34533838
- PMCID: PMC8726744
- DOI: 10.1002/prot.26237
Critical assessment of methods of protein structure prediction (CASP)-Round XIV
Abstract
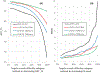
Critical assessment of structure prediction (CASP) is a community experiment to advance methods of computing three-dimensional protein structure from amino acid sequence. Core components are rigorous blind testing of methods and evaluation of the results by independent assessors. In the most recent experiment (CASP14), deep-learning methods from one research group consistently delivered computed structures rivaling the corresponding experimental ones in accuracy. In this sense, the results represent a solution to the classical protein-folding problem, at least for single proteins. The models have already been shown to be capable of providing solutions for problematic crystal structures, and there are broad implications for the rest of structural biology. Other research groups also substantially improved performance. Here, we describe these results and outline some of the many implications. Other related areas of CASP, including modeling of protein complexes, structure refinement, estimation of model accuracy, and prediction of inter-residue contacts and distances, are also described.
Keywords: Alphafold; CASP; community wide experiment; protein folding; protein structure prediction.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Figures

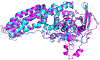



References
-
- Lensink MF, Brysbaert G, Mauri T, Nadzirin N, Velankar S, Chaleil RAG, Clarence T, Bates PA, Kong R, Liu B, Yang G, Liu M, Shi H, Lu X, Chang S, Roy RS, Quadir F, Liu J, Cheng J, Antoniak A, Czaplewski C, Gieldo NA, Kogut M, Lipska AG, Liwo A, Lubecka EA, Maszota-Zieleniak M, Sieradzan AK, Slusarz R, Wesolowski PA, ZiEba K, Del Carpio Munoz CA, Ichiishi E, Harmalkar A, Gray JJ, Bonvin A, Ambrosetti F, Honorato RV, Jandova Z, Jimenez-Garcia B, Koukos PI, Van Keulen S, Van Noort CW, Reau M, Roel-Touris J, Kotelnikov S, Padhorny D, Porter KA, Alekseenko A, Ignatov M, Desta I, Ashizawa R, Sun Z, Ghani U, Hashemi N, Vajda S, Kozakov D, Rosell M, Rodriguez-Lumbreras LA, Fernandez-Recio J, Karczynska A, Grudinin S, Yan Y, Li H, Lin P, Huang SY, Christoffer C, Terashi G, Verburgt J, Sarkar D, Aderinwale T, Wang X, Kihara D, Nakamura T, Hanazono Y, Gowthaman R, Guest JD, Yin R, Taherzadeh G, Pierce BG, Barradas-Bautista D, Cao Z, Cavallo L, Oliva R, Sun Y, Zhu S, Shen Y, Park T, Woo H, Yang J, Kwon S, Won J, Seok C, Kiyota Y, Kobayashi S, Harada Y, Takeda-Shitaka M, Kundrotas PJ, Singh A, Vakser IA, DapkUnas J, Olechnovic K, Venclovas C, Duan R, Qiu L, Zhang S, Zou X, Wodak SJ. Prediction of protein assemblies, the next frontier: The CASP14-CAPRI experiment. Proteins 2021. doi: 10.1002/prot.26222. Online ahead of print. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

