Plastome Structural Evolution and Homoplastic Inversions in Neo-Astragalus (Fabaceae)
- PMID: 34534296
- PMCID: PMC8486006
- DOI: 10.1093/gbe/evab215
Plastome Structural Evolution and Homoplastic Inversions in Neo-Astragalus (Fabaceae)
Abstract
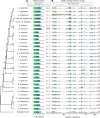
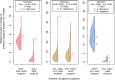
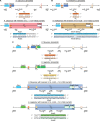
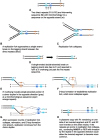
The plastid genomes of photosynthetic green plants have largely maintained conserved gene content and order as well as structure over hundreds of millions of years of evolution. Several plant lineages, however, have departed from this conservation and contain many plastome structural rearrangements, which have been associated with an abundance of repeated sequences both overall and near rearrangement endpoints. We sequenced the plastomes of 25 taxa of Astragalus L. (Fabaceae), a large genus in the inverted repeat-lacking clade of legumes, to gain a greater understanding of the connection between repeats and plastome inversions. We found plastome repeat structure has a strong phylogenetic signal among these closely related taxa mostly in the New World clade of Astragalus called Neo-Astragalus. Taxa without inversions also do not differ substantially in their overall repeat structure from four taxa each with one large-scale inversion. For two taxa with inversion endpoints between the same pairs of genes, differences in their exact endpoints indicate the inversions occurred independently. Our proposed mechanism for inversion formation suggests the short inverted repeats now found near the endpoints of the four inversions may be there as a result of these inversions rather than their cause. The longer inverted repeats now near endpoints may have allowed the inversions first mediated by shorter microhomologous sequences to propagate, something that should be considered in explaining how any plastome rearrangement becomes fixed regardless of the mechanism of initial formation.
Keywords: chloroplast; inverted repeat-lacking clade; legumes; microhomology-mediated rearrangements; plastid genome.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. - PubMed
-
- Azani N, Bruneau A, Wojciechowski MF, Zarre S.. 2019. Miocene climate change as a driving force for multiple origins of annual species in Astragalus (Fabaceae, Papilionoideae). Mol Phylogenet Evol. 137:210–221. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

