COPII Sec23 proteins form isoform-specific endoplasmic reticulum exit sites with differential effects on polarized growth
- PMID: 34534343
- PMCID: PMC8846183
- DOI: 10.1093/plcell/koab229
COPII Sec23 proteins form isoform-specific endoplasmic reticulum exit sites with differential effects on polarized growth
Abstract
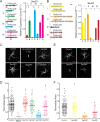
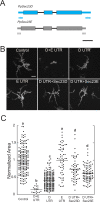
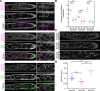
Coat Protein complex II (COPII), a coat protein complex that forms vesicles on the endoplasmic reticulum (ER), mediates trafficking to the Golgi. While metazoans have few genes encoding each COPII component, plants have expanded these gene families, leading to the hypothesis that plant COPII has functionally diversified. In the moss Physcomitrium (Physcomitrella) patens, the Sec23/24 gene families are each composed of seven genes. Silencing Sec23/24 revealed isoform-specific contributions to polarized growth, with the closely related Sec23D/E and Sec24C/D essential for protonemal development. Focusing on Sec23, we discovered that Sec23D/E mediate ER-to Golgi transport and are essential for tip growth, with Sec23D localizing to presumptive ER exit sites. In contrast, Sec23A, B, C, F, and G are dispensable and do not quantitatively affect ER-to-Golgi trafficking. However, Δsec23abcfg plants exhibited reduced secretion of plasma membrane cargo. Of the four highly expressed protonemal Sec23 genes, Sec23F/G are members of a divergent Sec23 clade specifically retained in land plants. Notably, Sec23G accumulates on ER-associated foci that are significantly larger, do not overlap with, and are independent of Sec23D. While Sec23D/E form ER exit sites and function as bona fide COPII components essential for tip-growing protonemata, Sec23G and the closely related Sec23F have likely functionally diversified, forming separate and independent ER exit sites and participating in Golgi-independent trafficking pathways.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures





Comment in
-
Back to the roots: A focus on plant cell biology.Plant Cell. 2022 Jan 20;34(1):1-3. doi: 10.1093/plcell/koab278. Plant Cell. 2022. PMID: 34755878 Free PMC article. No abstract available.
References
-
- Aridor M (2018) COPII gets in shape: lessons derived from morphological aspects of early secretion. Traffic 19: 823–839 - PubMed
-
- Barlow LD, Dacks JB (2018) Seeing the endomembrane system for the trees: evolutionary analysis highlights the importance of plants as models for eukaryotic membrane-trafficking. Semin Cell Dev Biol 80: 142–152 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

