Methods in sulfide and persulfide research
- PMID: 34534626
- PMCID: PMC8486624
- DOI: 10.1016/j.niox.2021.09.002
Methods in sulfide and persulfide research
Abstract
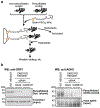
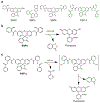
Sulfides and persulfides/polysulfides (R-Sn-R', n > 2; R-Sn-H, n > 1) are endogenously produced metabolites that are abundant in mammalian and human cells and tissues. The most typical persulfides that are widely distributed among different organisms include various reactive persulfides-low-molecular-weight thiol compounds such as cysteine hydropersulfide, glutathione hydropersulfide, and glutathione trisulfide as well as protein-bound thiols. These species are generally more redox-active than are other simple thiols and disulfides. Although hydrogen sulfide (H2S) has been suggested for years to be a small signaling molecule, it is intimately linked biochemically to persulfides and may actually be more relevant as a marker of functionally active persulfides. Reactive persulfides can act as powerful antioxidants and redox signaling species and are involved in energy metabolism. Recent evidence revealed that cysteinyl-tRNA synthetases (CARSs) act as the principal cysteine persulfide synthases in mammals and contribute significantly to endogenous persulfide/polysulfide production, in addition to being associated with a battery of enzymes including cystathionine β-synthase, cystathionine γ-lyase, and 3-mercaptopyruvate sulfurtransferase, which have been described as H2S-producing enzymes. The reactive sulfur metabolites including persulfides/polysulfides derived from CARS2, a mitochondrial isoform of CARS, also mediate not only mitochondrial biogenesis and bioenergetics but also anti-inflammatory and immunomodulatory functions. The physiological roles of persulfides, their biosynthetic pathways, and their pathophysiology in various diseases are not fully understood, however. Developing basic and high precision techniques and methods for the detection, characterization, and quantitation of sulfides and persulfides is therefore of great importance so as to thoroughly understand and clarify the exact functions and roles of these species in cells and in vivo.
Keywords: Hydrogen sulfide; Methodology; Persulfides/polysulfides; Reactive sulfur metabolites; Sulfides.
Copyright © 2021 Elsevier Inc. All rights reserved.
Figures








Similar articles
-
Synthesis of Sulfides and Persulfides Is Not Impeded by Disruption of Three Canonical Enzymes in Sulfur Metabolism.Antioxidants (Basel). 2023 Apr 3;12(4):868. doi: 10.3390/antiox12040868. Antioxidants (Basel). 2023. PMID: 37107243 Free PMC article.
-
Enzymatic Regulation and Biological Functions of Reactive Cysteine Persulfides and Polysulfides.Biomolecules. 2020 Aug 27;10(9):1245. doi: 10.3390/biom10091245. Biomolecules. 2020. PMID: 32867265 Free PMC article. Review.
-
Persulfide synthases that are functionally coupled with translation mediate sulfur respiration in mammalian cells.Br J Pharmacol. 2019 Feb;176(4):607-615. doi: 10.1111/bph.14356. Epub 2018 Jun 7. Br J Pharmacol. 2019. PMID: 29748969 Free PMC article. Review.
-
Reactive Sulfur Species: A New Redox Player in Cardiovascular Pathophysiology.Arterioscler Thromb Vasc Biol. 2020 Apr;40(4):874-884. doi: 10.1161/ATVBAHA.120.314084. Epub 2020 Mar 5. Arterioscler Thromb Vasc Biol. 2020. PMID: 32131614 Free PMC article.
-
Persulfides, at the crossroads between hydrogen sulfide and thiols.Essays Biochem. 2020 Feb 17;64(1):155-168. doi: 10.1042/EBC20190049. Essays Biochem. 2020. PMID: 32016341 Review.
Cited by
-
A Biochemical Corrosion Monitoring Sensor with a Silver/Carbon Comb Structure for the Detection of Living Escherichia coli.ACS Omega. 2023 Nov 13;8(46):43511-43520. doi: 10.1021/acsomega.3c03632. eCollection 2023 Nov 21. ACS Omega. 2023. PMID: 38027348 Free PMC article.
-
Hydropersulfides (RSSH) Outperform Post-Conditioning and Other Reactive Sulfur Species in Limiting Ischemia-Reperfusion Injury in the Isolated Mouse Heart.Antioxidants (Basel). 2022 May 20;11(5):1010. doi: 10.3390/antiox11051010. Antioxidants (Basel). 2022. PMID: 35624878 Free PMC article.
-
Supersulfide metabolome of exhaled breath condensate applied as diagnostic biomarkers for esophageal cancer.Cancer Sci. 2025 Apr;116(4):1023-1033. doi: 10.1111/cas.16430. Epub 2025 Feb 2. Cancer Sci. 2025. PMID: 39895210 Free PMC article.
-
The Therapeutic Potential of Supersulfides in Oxidative Stress-Related Diseases.Biomolecules. 2025 Jan 23;15(2):172. doi: 10.3390/biom15020172. Biomolecules. 2025. PMID: 40001475 Free PMC article. Review.
-
Supersulfide formation in the sinus mucosa of chronic rhinosinusitis.Laryngoscope Investig Otolaryngol. 2024 Jul 27;9(4):e1261. doi: 10.1002/lio2.1261. eCollection 2024 Aug. Laryngoscope Investig Otolaryngol. 2024. PMID: 39071205 Free PMC article.
References
-
- Pryor WA, Houk KN, Foote CS, Fukuto JM, Ignarro LJ, Squadrito GL, Davies KJA, Free radical biology and medicine, it’s a gas, man, Am. J. Physiol. Regul. Integr. Comp. Physiol 291 (2006) R491–R511. - PubMed
-
- Fukuto JM, Carrington SJ, Tantillo DJ, Harrison JG, Ignarro LJ, Freeman BA, Chen A, Wink DA, Small molecule signaling agents: the integrated chemistry and biochemistry of nitrogen oxides, oxides of carbon, dioxygen, hydrogen sulfide, and their derived species, Chem. Res. Toxicol 25 (2012) 769–793. - PMC - PubMed
-
- DeMartino AW, Zigler DF, Fukuto JM, Ford PC, Carbon disulfide. Just toxic or also bioregulatory and/or therapeutic? Chem. Soc. Rev 46 (2017) 21–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

