Adipocytes disrupt the translational programme of acute lymphoblastic leukaemia to favour tumour survival and persistence
- PMID: 34535653
- PMCID: PMC8448863
- DOI: 10.1038/s41467-021-25540-4
Adipocytes disrupt the translational programme of acute lymphoblastic leukaemia to favour tumour survival and persistence
Abstract
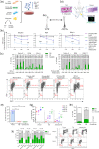
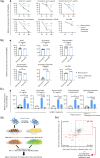
The specific niche adaptations that facilitate primary disease and Acute Lymphoblastic Leukaemia (ALL) survival after induction chemotherapy remain unclear. Here, we show that Bone Marrow (BM) adipocytes dynamically evolve during ALL pathogenesis and therapy, transitioning from cellular depletion in the primary leukaemia niche to a fully reconstituted state upon remission induction. Functionally, adipocyte niches elicit a fate switch in ALL cells towards slow-proliferation and cellular quiescence, highlighting the critical contribution of the adipocyte dynamic to disease establishment and chemotherapy resistance. Mechanistically, adipocyte niche interaction targets posttranscriptional networks and suppresses protein biosynthesis in ALL cells. Treatment with general control nonderepressible 2 inhibitor (GCN2ib) alleviates adipocyte-mediated translational repression and rescues ALL cell quiescence thereby significantly reducing the cytoprotective effect of adipocytes against chemotherapy and other extrinsic stressors. These data establish how adipocyte driven restrictions of the ALL proteome benefit ALL tumours, preventing their elimination, and suggest ways to manipulate adipocyte-mediated ALL resistance.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Goldstone AH, et al. In adults with standard-risk acute lymphoblastic leukemia, the greatest benefit is achieved from a matched sibling allogeneic transplantation in first complete remission, and an autologous transplantation is less effective than conventional consolidation/maintenance chemotherapy in all patients: final results of the International ALL Trial (MRC UKALL XII/ECOG E2993) Blood. 2008;111:1827–1833. doi: 10.1182/blood-2007-10-116582. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

