Impacts of environmental conditions, and allelic variation of cytosolic glutamine synthetase on maize hybrid kernel production
- PMID: 34535763
- PMCID: PMC8448750
- DOI: 10.1038/s42003-021-02598-w
Impacts of environmental conditions, and allelic variation of cytosolic glutamine synthetase on maize hybrid kernel production
Abstract
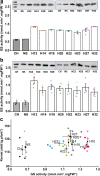
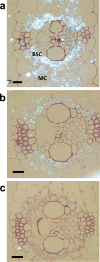
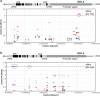
Cytosolic glutamine synthetase (GS1) is the enzyme mainly responsible of ammonium assimilation and reassimilation in maize leaves. The agronomic potential of GS1 in maize kernel production was investigated by examining the impact of an overexpression of the enzyme in the leaf cells. Transgenic hybrids exhibiting a three-fold increase in leaf GS activity were produced and characterized using plants grown in the field. Several independent hybrids overexpressing Gln1-3, a gene encoding cytosolic (GS1), in the leaf and bundle sheath mesophyll cells were grown over five years in different locations. On average, a 3.8% increase in kernel yield was obtained in the transgenic hybrids compared to controls. However, we observed that such an increase was simultaneously dependent upon both the environmental conditions and the transgenic event for a given field trial. Although variable from one environment to another, significant associations were also found between two GS1 genes (Gln1-3 and Gln1-4) polymorphic regions and kernel yield in different locations. We propose that the GS1 enzyme is a potential lead for producing high yielding maize hybrids using either genetic engineering or marker-assisted selection. However, for these hybrids, yield increases will be largely dependent upon the environmental conditions used to grow the plants.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Hirel, B. & Lea, P. J. in The Zea mays Genome. Compendium of Plant Genomes (eds. Bennetzen, J., Flint-Garcia, S. Hirsch, C. & Tuberosa, R.) 259–286 (Springer International Publishing, 2018).
-
- Hirel, B. & Lea, P. J. in The Molecular and Physiological Basis of Nutrient Use Efficiency in Crops (eds. Hawkesford, M. J. & Barraclough, P. B.) 139–164 (Wiley-Blackwell, 2011).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

