Resolving distance variations by single-molecule FRET and EPR spectroscopy using rotamer libraries
- PMID: 34536387
- PMCID: PMC8595751
- DOI: 10.1016/j.bpj.2021.09.021
Resolving distance variations by single-molecule FRET and EPR spectroscopy using rotamer libraries
Abstract
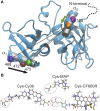
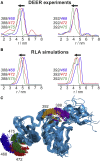
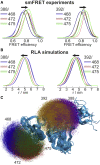
Förster resonance energy transfer (FRET) and electron paramagnetic resonance (EPR) spectroscopy are complementary techniques for quantifying distances in the nanometer range. Both approaches are commonly employed for probing the conformations and conformational changes of biological macromolecules based on site-directed fluorescent or paramagnetic labeling. FRET can be applied in solution at ambient temperature and thus provides direct access to dynamics, especially if used at the single-molecule level, whereas EPR requires immobilization or work at cryogenic temperatures but provides data that can be more reliably used to extract distance distributions. However, a combined analysis of the complementary data from the two techniques has been complicated by the lack of a common modeling framework. Here, we demonstrate a systematic analysis approach based on rotamer libraries for both FRET and EPR labels to predict distance distributions between two labels from a structural model. Dynamics of the fluorophores within these distance distributions are taken into account by diffusional averaging, which improves the agreement with experiment. Benchmarking this methodology with a series of surface-exposed pairs of sites in a structured protein domain reveals that the lowest resolved distance differences can be as small as ∼0.25 nm for both techniques, with quantitative agreement between experimental and simulated transfer efficiencies within a range of ±0.045. Rotamer library analysis thus establishes a coherent way of treating experimental data from EPR and FRET and provides a basis for integrative structural modeling, including studies of conformational distributions and dynamics of biological macromolecules using both techniques.
Copyright © 2021 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
When two become one: Integrating FRET and EPR into one structural model.Biophys J. 2021 Nov 2;120(21):4637-4638. doi: 10.1016/j.bpj.2021.09.022. Epub 2021 Sep 16. Biophys J. 2021. PMID: 34555323 Free PMC article. No abstract available.
References
-
- Schlundt A., Tants J.-N., Sattler M. Integrated structural biology to unravel molecular mechanisms of protein-RNA recognition. Methods. 2017;118–119:119–136. - PubMed
-
- Dimitrova-Paternoga L., Jagtap P.K.A., et al. Hennig J. Integrative structural biology of protein-RNA complexes. Structure. 2020;28:6–28. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

