The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages
- PMID: 34537136
- PMCID: PMC8421097
- DOI: 10.1016/j.cell.2021.09.003
The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages
Abstract
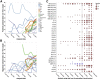
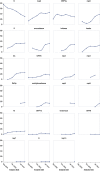
The independent emergence late in 2020 of the B.1.1.7, B.1.351, and P.1 lineages of SARS-CoV-2 prompted renewed concerns about the evolutionary capacity of this virus to overcome public health interventions and rising population immunity. Here, by examining patterns of synonymous and non-synonymous mutations that have accumulated in SARS-CoV-2 genomes since the pandemic began, we find that the emergence of these three "501Y lineages" coincided with a major global shift in the selective forces acting on various SARS-CoV-2 genes. Following their emergence, the adaptive evolution of 501Y lineage viruses has involved repeated selectively favored convergent mutations at 35 genome sites, mutations we refer to as the 501Y meta-signature. The ongoing convergence of viruses in many other lineages on this meta-signature suggests that it includes multiple mutation combinations capable of promoting the persistence of diverse SARS-CoV-2 lineages in the face of mounting host immune recognition.
Keywords: COVID 19; convergent mutations; directional selection; diversifying selection; evolutionary adaptation; immune evasion; lineage-defining mutations; positive selection; recurrent mutations; transmission advantage.
Copyright © 2021. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests J.O.W. has been funded by Gilead Sciences, LLC (completed) and the CDC (ongoing) via grants and contracts to his institution unrelated to this research.
Figures






Update of
-
The emergence and ongoing convergent evolution of the N501Y lineages coincides with a major global shift in the SARS-CoV-2 selective landscape.medRxiv [Preprint]. 2021 Jul 25:2021.02.23.21252268. doi: 10.1101/2021.02.23.21252268. medRxiv. 2021. Update in: Cell. 2021 Sep 30;184(20):5189-5200.e7. doi: 10.1016/j.cell.2021.09.003. PMID: 33688681 Free PMC article. Updated. Preprint.
References
-
- Althaus C.L., Baggio S., Reichmuth M.L., Hodcroft E.B., Riou J., Neher R.A., Jacquerioz F., Spechbach H., Salamun J., Vetter P., et al. A tale of two variants: Spread of SARS-CoV-2 variants Alpha in Geneva, Switzerland, and Beta in South Africa. MedRxiv. 2021 doi: 10.1101/2021.02.23.21252268. - DOI
-
- Annavajhala M.K., Mohri H., Zucker J.E., Sheng Z., Wang P., Gomez-Simmonds A., Ho D.D., Uhlemann A.-C. A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. MedRxiv. 2021 doi: 10.1101/2021.02.23.21252259. - DOI
-
- Campbell K.M., Steiner G., Wells D.K., Ribas A., Kalbasi A. Prediction of SARS-CoV-2 epitopes across 9360 HLA class I alleles. BioRxiv. 2020 doi: 10.1101/2020.03.30.016931. - DOI
-
- Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Network for Genomic Surveillance in South Africa. COMMIT-KZN Team Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. - PMC - PubMed
-
- Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., et al. CITIID-NIHR BioResource COVID-19 Collaboration. COVID-19 Genomics UK (COG-UK) Consortium Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

