Genome-wide detection and classification of terpene synthase genes in Aquilaria agallochum
- PMID: 34539112
- PMCID: PMC8405786
- DOI: 10.1007/s12298-021-01040-z
Genome-wide detection and classification of terpene synthase genes in Aquilaria agallochum
Abstract
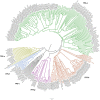
Agarwood, one of the precious woods in the globe, is produced by Aquilaria plant species during an upshot of wounding and infection. Produced as a defence response, the dark, fragrant resin gets secreted in the plant's duramen, which is impregnated with fragrant molecules with the due course. Agarwood has gained worldwide popularity due to its high aromatic oil, fragrance, and pharmaceutical value, which makes it highly solicited by numerous industries. Predominant chemical constituents of agarwood, sesquiterpenoids, and 2-(2-phenylethyl) chromones have been scrutinized to comprehend the scientific nature of the fragrant wood and develop novel products. However, the genes involved in the biosynthesis of these aromatic compounds are still not comprehensively studied in Aquilaria. In this study, publicly available genomic and transcriptomics data of Aquilaria agallochum were integrated to identify putative functional terpene synthase genes (TPSs). The in silico study enabled us to identify ninety-six TPSs, of which thirty-nine full-length genes were systematically classified into TPS-a, TPS-b, TPS-c, TPS-e, TPS-f, and TPS-g subfamilies based on their gene structure, conserve motif, and phylogenetic comparison with TPSs from other plant species. Analysis of the cis-regulatory elements present upstream of AaTPSs revealed their association with hormone, stress and light responses. In silico expression studies detected their up-regulation in stress induced tissue. This study provides a basic understanding of terpene synthase gene repertoire in Aquilaria agallochum and unlatches opportunities for the biochemical characterization and biotechnological exploration of these genes.
Supplementary information: The online version contains supplementary material available at 10.1007/s12298-021-01040-z.
Keywords: Aquilaria agallochum; Classification; Phylogenetic analysis and functional prediction; Terpene synthase.
© Prof. H.S. Srivastava Foundation for Science and Society 2021.
Conflict of interest statement
Conflict of interestThe authors declares no conflict of interest exist.
Figures






References
-
- Akter S, Islam MT, Zulkefeli M, Khan SI. Agarwood production-a multidisciplinary field to be explored in Bangladesh. Int J Pharm Life Sci. 2013;2:22–32. doi: 10.3329/ijpls.v2i1.15132. - DOI
-
- Azzarina AB, Mohamed R, Lee SY, Nazre M. Temporal and spatial expression of terpene synthase genes associated with agarwood formation in Aquilaria malaccensis Lam. N Z J Sci. 2016;46:12. doi: 10.1186/s40490-016-0068-9. - DOI
LinkOut - more resources
Full Text Sources
