Cultivation of Important Methanotrophs From Indian Rice Fields
- PMID: 34539593
- PMCID: PMC8447245
- DOI: 10.3389/fmicb.2021.669244
Cultivation of Important Methanotrophs From Indian Rice Fields
Abstract
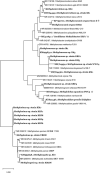
Methanotrophs are aerobic to micro-aerophilic bacteria, which oxidize and utilize methane, the second most important greenhouse gas. The community structure of the methanotrophs in rice fields worldwide has been studied mainly using culture-independent methods. Very few studies have focused on culturing methanotrophs from rice fields. We developed a unique method for the cultivation of methanotrophs from rice field samples. Here, we used a modified dilute nitrate mineral salts (dNMS) medium, with two cycles of dilution till extinction series cultivation with prolonged incubation time, and used agarose in the solid medium. The cultivation approach resulted in the isolation of methanotrophs from seven genera from the three major groups: Type Ia (Methylomonas, Methylomicrobium, and Methylocucumis), Type Ib (Methylocaldum and Methylomagnum), and Type II (Methylocystis and Methylosinus). Growth was obtained till 10-6-10-8 dilutions in the first dilution series, indicating the culturing of dominant methanotrophs. Our study was supported by 16S rRNA gene-based next-generation sequencing (NGS) of three of the rice samples. Our analyses and comparison with the global scenario suggested that the cultured members represented the major detected taxa. Strain RS1, representing a putative novel species of Methylomicrobium, was cultured; and the draft genome sequence was obtained. Genome analysis indicated that RS1 represented a new putative Methylomicrobium species. Methylomicrobium has been detected globally in rice fields as a dominant genus, although no Methylomicrobium strains have been isolated from rice fields worldwide. Ours is one of the first extensive studies on cultured methanotrophs from Indian rice fields focusing on the tropical region, and a unique method was developed. A total of 29 strains were obtained, which could be used as models for studying methane mitigation from rice fields and for environmental and biotechnological applications.
Keywords: India; cultivation; metagenomics; methane; methanotrophs; novel; rice fields; serial dilution.
Copyright © 2021 Rahalkar, Khatri, Pandit, Bahulikar and Mohite.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Antony C. P., Doronina N. V., Boden R., Trotsenko Y. A., Shouche Y. S., Murrell J. C. (2012). Methylophaga lonarensis sp. nov., a moderately haloalkaliphilic methylotroph isolated from the soda lake sediments of a meteorite impact crater. Int. J. Syst. Evolut. Microbiol. 62 1613–1618. 10.1099/ijs.0.035089-0 - DOI - PubMed
-
- Bowman J. (2006). The methanotrophs—the families methylococcaceae and methylocystaceae. Prokaryotes 5 266–289. 10.1007/0-387-30745-1_15 - DOI
-
- Bowman J. (2011). “Approaches for the characterization and description of novel methanotrophic bacteria,” in Methods in Methane Metabolism, Part B Methanotrophy Methods in Enzymology, eds Rosenzweig A. C., Ragsdale S. W. (Amsterdam: Elsevier; ), 45–62. 10.1016/b978-0-12-386905-0.00004-8 - DOI - PubMed
LinkOut - more resources
Full Text Sources

