Allele-specific assembly of a eukaryotic genome corrects apparent frameshifts and reveals a lack of nonsense-mediated mRNA decay
- PMID: 34541528
- PMCID: PMC8445201
- DOI: 10.1093/nargab/lqab082
Allele-specific assembly of a eukaryotic genome corrects apparent frameshifts and reveals a lack of nonsense-mediated mRNA decay
Abstract
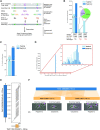
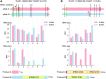
To date, most reference genomes represent a mosaic consensus sequence in which the homologous chromosomes are collapsed into one sequence. This approach produces sequence artefacts and impedes analyses of allele-specific mechanisms. Here, we report an allele-specific genome assembly of the diploid parasite Trypanosoma brucei and reveal allelic variants affecting gene expression. Using long-read sequencing and chromosome conformation capture data, we could assign 99.5% of all heterozygote variants to a specific homologous chromosome and build a 66 Mb long allele-specific genome assembly. The phasing of haplotypes allowed us to resolve hundreds of artefacts present in the previous mosaic consensus assembly. In addition, it revealed allelic recombination events, visible as regions of low allelic heterozygosity, enabling the lineage tracing of T. brucei isolates. Interestingly, analyses of transcriptome and translatome data of genes with allele-specific premature termination codons point to the absence of a nonsense-mediated decay mechanism in trypanosomes. Taken together, this study delivers a reference quality allele-specific genome assembly of T. brucei and demonstrates the importance of such assemblies for the study of gene expression control. We expect the new genome assembly will increase the awareness of allele-specific phenomena and provide a platform to investigate them.
© The Author(s) 2021. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures




References
-
- Bertelli C., Greub G. Rapid bacterial genome sequencing: methods and applications in clinical microbiology. Clin. Microbiol. Infect. 2013; 19:803–813. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

