Salivary Metabolites are Promising Non-Invasive Biomarkers of Hepatocellular Carcinoma and Chronic Liver Disease
- PMID: 34541549
- PMCID: PMC8447405
- DOI: 10.1002/lci2.25
Salivary Metabolites are Promising Non-Invasive Biomarkers of Hepatocellular Carcinoma and Chronic Liver Disease
Abstract
Background: Hepatocellular carcinoma (HCC) is a leading causes of cancer mortality worldwide. Improved tools are needed for detecting HCC so that treatment can begin as early as possible. Current diagnostic approaches and existing biomarkers, such as alpha-fetoprotein (AFP) lack sensitivity, resulting in too many false negative diagnoses. Machine-learning may be able to identify combinations of biomarkers that provide more robust predictions and improve sensitivity for detecting HCC. We sought to evaluate whether metabolites in patient saliva could distinguish those with HCC, cirrhosis, and those with no documented liver disease.
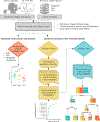
Methods and results: We tested 125 salivary metabolites from 110 individuals (43 healthy, 37 HCC, 30 cirrhosis) and identified 4 metabolites that displayed significantly different abundance between groups (FDR P <.2). We also developed four tree-based, machine-learning models, optimized to include different numbers of metabolites, that were trained using cross-validation on 99 patients and validated on a withheld test set of 11 patients. A model using 12 metabolites -octadecanol, acetophenone, lauric acid, 1-monopalmitin, dodecanol, salicylaldehyde, glycyl-proline, 1-monostearin, creatinine, glutamine, serine and 4-hydroxybutyric acid- had a cross-validated sensitivity of 84.8%, specificity of 92.4% and correctly classified 90% of the HCC patients in the test cohort. This model outperformed previously reported sensitivities and specificities for AFP (20-100ng/ml) (61%, 86%) and AFP plus ultrasound (62%, 88%).
Conclusions and impact: Metabolites detectable in saliva may represent products of disease pathology or a breakdown in liver function. Notably, combinations of salivary metabolites derived from machine-learning may serve as promising non-invasive biomarkers for the detection of HCC.
Keywords: Metabolomics; cirrhosis; liver cancer; machine learnings; risk factor.
Conflict of interest statement
Competing Interests D.M.R. has an equity stake in Interpares Biomedicine, LLC. D.M.R., F.A., D.S.A hold intellectual property related to the detection of hepatocellular carcinoma.
Figures

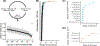

References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. United States; 2020;70:7–30. - PubMed
-
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet Lond Engl. England; 2018;391:1301–14. - PubMed
-
- Sun C-A, Wu D-M, Lin C-C, Lu S-N, You S-L, Wang L-Y, et al. Incidence and cofactors of hepatitis C virus-related hepatocellular carcinoma: a prospective study of 12,008 men in Taiwan. Am J Epidemiol. United States; 2003;157:674–82. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
