Delete and survive: strategies of programmed genetic material elimination in eukaryotes
- PMID: 34542224
- PMCID: PMC9292451
- DOI: 10.1111/brv.12796
Delete and survive: strategies of programmed genetic material elimination in eukaryotes
Abstract
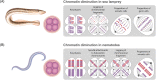
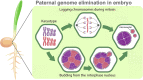
Genome stability is a crucial feature of eukaryotic organisms because its alteration drastically affects the normal development and survival of cells and the organism as a whole. Nevertheless, some organisms can selectively eliminate part of their genomes from certain cell types during specific stages of ontogenesis. This review aims to describe the phenomenon of programmed DNA elimination, which includes chromatin diminution (together with programmed genome rearrangement or DNA rearrangements), B and sex chromosome elimination, paternal genome elimination, parasitically induced genome elimination, and genome elimination in animal and plant hybrids. During programmed DNA elimination, individual chromosomal fragments, whole chromosomes, and even entire parental genomes can be selectively removed. Programmed DNA elimination occurs independently in different organisms, ranging from ciliate protozoa to mammals. Depending on the sequences destined for exclusion, programmed DNA elimination may serve as a radical mechanism of dosage compensation and inactivation of unnecessary or dangerous genetic entities. In hybrids, genome elimination results from competition between parental genomes. Despite the different consequences of DNA elimination, all genetic material destined for elimination must be first recognised, epigenetically marked, separated, and then removed and degraded.
Keywords: B chromosomes; asexual hybrids; chromatin diminution; chromosomal lagging; micronuclei; programmed DNA elimination; sex chromosomes.
© 2021 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
Figures






References
-
- Adams, R. R. , Maiato, H. , Earnshaw, W. C. & Carmena, M. (2001). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. The Journal of Cell Biology 153, 865–880. - PMC - PubMed
-
- Akifyev, A. P. & Grishanin, A. K. (2005). Some conclusions on the role of redundant DNA and the mechanisms of eukaryotic genome evolution inferred from studies of chromatin diminution in Cyclopoida. Russian Journal of Genetics 41, 366–377. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

