SMA-miRs (miR-181a-5p, -324-5p, and -451a) are overexpressed in spinal muscular atrophy skeletal muscle and serum samples
- PMID: 34542403
- PMCID: PMC8486378
- DOI: 10.7554/eLife.68054
SMA-miRs (miR-181a-5p, -324-5p, and -451a) are overexpressed in spinal muscular atrophy skeletal muscle and serum samples
Abstract
Background: Spinal muscular atrophy (SMA) is a neuromuscular disorder characterized by the degeneration of the second motor neuron. The phenotype ranges from very severe to very mild forms. All patients have the homozygous loss of the SMN1 gene and a variable number of SMN2 (generally 2-4 copies), inversely related to the severity. The amazing results of the available treatments have made compelling the need of prognostic biomarkers to predict the progression trajectories of patients. Besides the SMN2 products, few other biomarkers have been evaluated so far, including some miRs.
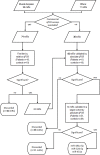
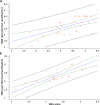
Methods: We performed whole miRNome analysis of muscle samples of patients and controls (14 biopsies and 9 cultures). The levels of muscle differentially expressed miRs were evaluated in serum samples (51 patients and 37 controls) and integrated with SMN2 copies, SMN2 full-length transcript levels in blood and age (SMA-score).
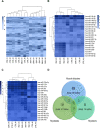
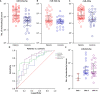
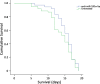
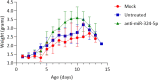
Results: Over 100 miRs were differentially expressed in SMA muscle; 3 of them (hsa-miR-181a-5p, -324-5p, -451a; SMA-miRs) were significantly upregulated in the serum of patients. The severity predicted by the SMA-score was related to that of the clinical classification at a correlation coefficient of 0.87 (p<10-5).
Conclusions: miRNome analyses suggest the primary involvement of skeletal muscle in SMA pathogenesis. The SMA-miRs are likely actively released in the blood flow; their function and target cells require to be elucidated. The accuracy of the SMA-score needs to be verified in replicative studies: if confirmed, its use could be crucial for the routine prognostic assessment, also in presymptomatic patients.
Funding: Telethon Italia (grant #GGP12116).
Keywords: SMN1; biomarker; genetics; genomics; human; mRNA; medicine; miRNA; mouse; skeletal muscle; spinal muscular atrophy.
© 2021, Abiusi et al.
Conflict of interest statement
EA, PI, CC, LL, MP, GC, MP, AD, FD, AN, SS, SF, GB, IM, MM, MP, KP, LL, LT, FR, CB, DL, EB, LM, EM, LD, FT No competing interests declared, DD Davide D'Amico is affiliated with Amazentis SA. The author has no financial interests to declare. At the time of study developement, Dr. D'Amico had an academic affiliation
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

