Condensation tendency and planar isotropic actin gradient induce radial alignment in confined monolayers
- PMID: 34542405
- PMCID: PMC8478414
- DOI: 10.7554/eLife.60381
Condensation tendency and planar isotropic actin gradient induce radial alignment in confined monolayers
Abstract
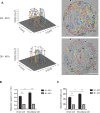
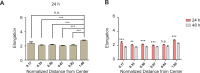
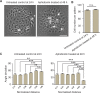
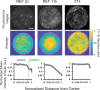
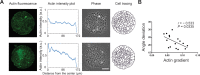
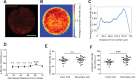
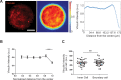
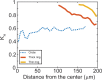
A monolayer of highly motile cells can establish long-range orientational order, which can be explained by hydrodynamic theory of active gels and fluids. However, it is less clear how cell shape changes and rearrangement are governed when the monolayer is in mechanical equilibrium states when cell motility diminishes. In this work, we report that rat embryonic fibroblasts (REF), when confined in circular mesoscale patterns on rigid substrates, can transition from the spindle shapes to more compact morphologies. Cells align radially only at the pattern boundary when they are in the mechanical equilibrium. This radial alignment disappears when cell contractility or cell-cell adhesion is reduced. Unlike monolayers of spindle-like cells such as NIH-3T3 fibroblasts with minimal intercellular interactions or epithelial cells like Madin-Darby canine kidney (MDCK) with strong cortical actin network, confined REF monolayers present an actin gradient with isotropic meshwork, suggesting the existence of a stiffness gradient. In addition, the REF cells tend to condense on soft substrates, a collective cell behavior we refer to as the 'condensation tendency'. This condensation tendency, together with geometrical confinement, induces tensile prestretch (i.e. an isotropic stretch that causes tissue to contract when released) to the confined monolayer. By developing a Voronoi-cell model, we demonstrate that the combined global tissue prestretch and cell stiffness differential between the inner and boundary cells can sufficiently define the cell radial alignment at the pattern boundary.
Keywords: actin gradient; contractility; fibroblast; microcontact printing; physics of living systems; prestretch; radial alignment; rat.
© 2021, Xie et al.
Conflict of interest statement
TX, SS, NO, LB, MW, YS No competing interests declared
Figures


















Similar articles
-
Cell-Cell Adhesion and Cortical Actin Bending Govern Cell Elongation on Negatively Curved Substrates.Biophys J. 2018 Apr 10;114(7):1707-1717. doi: 10.1016/j.bpj.2018.02.027. Biophys J. 2018. PMID: 29642039 Free PMC article.
-
Advancing Edge Speeds of Epithelial Monolayers Depend on Their Initial Confining Geometry.PLoS One. 2016 Apr 14;11(4):e0153471. doi: 10.1371/journal.pone.0153471. eCollection 2016. PLoS One. 2016. PMID: 27078632 Free PMC article.
-
Mechanical adaptions of collective cells nearby free tissue boundaries.J Biomech. 2020 May 7;104:109763. doi: 10.1016/j.jbiomech.2020.109763. Epub 2020 Mar 22. J Biomech. 2020. PMID: 32224050
-
On the mechanisms of cortical actin flow and its role in cytoskeletal organisation of fibroblasts.Symp Soc Exp Biol. 1993;47:35-56. Symp Soc Exp Biol. 1993. PMID: 8165576 Review.
-
Organization and polarity of actin filament networks in cells: implications for the mechanism of myosin-based cell motility.Biochem Soc Symp. 1999;65:173-205. Biochem Soc Symp. 1999. PMID: 10320939 Review.
Cited by
-
Tendon-associated gene expression precedes osteogenesis in mid-palatal suture establishment.bioRxiv [Preprint]. 2024 May 14:2024.05.11.590129. doi: 10.1101/2024.05.11.590129. bioRxiv. 2024. PMID: 38798531 Free PMC article. Preprint.
-
Multicellular aligned bands disrupt global collective cell behavior.Acta Biomater. 2023 Jun;163:117-130. doi: 10.1016/j.actbio.2022.10.041. Epub 2022 Oct 26. Acta Biomater. 2023. PMID: 36306982 Free PMC article.
References
-
- Aomatsu E, Chosa N, Nishihira S, Sugiyama Y, Miura H, Ishisaki A. Cell-cell adhesion through N-cadherin enhances VCAM-1 expression via PDGFRβ in a ligand-independent manner in mesenchymal stem cells. International Journal of Molecular Medicine. 2014;33:565–572. doi: 10.3892/ijmm.2013.1607. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

