Defining the transcriptional control of pediatric AML highlights RARA as a superenhancer-regulated druggable dependency
- PMID: 34543389
- PMCID: PMC9153032
- DOI: 10.1182/bloodadvances.2020003737
Defining the transcriptional control of pediatric AML highlights RARA as a superenhancer-regulated druggable dependency
Abstract
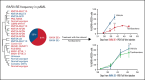
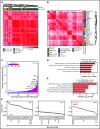
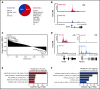
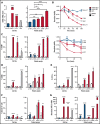
Somatic mutations are rare in pediatric acute myeloid leukemia (pAML), indicating that alternate strategies are needed to identify targetable dependencies. We performed the first enhancer mapping of pAML in 22 patient samples. Generally, pAML samples were distinct from adult AML samples, and MLL (KMT2A)-rearranged samples were also distinct from non-KMT2A-rearranged samples. Focusing specifically on superenhancers (SEs), we identified SEs associated with many known leukemia regulators. The retinoic acid receptor alpha (RARA) gene was differentially regulated in our cohort, and a RARA-associated SE was detected in 64% of the study cohort across all cytogenetic and molecular subtypes tested. RARA SE+ pAML cell lines and samples exhibited high RARA messenger RNA levels. These samples were specifically sensitive to the synthetic RARA agonist tamibarotene in vitro, with slowed proliferation, apoptosis induction, differentiation, and upregulated retinoid target gene expression, compared with RARA SE- samples. Tamibarotene prolonged survival and suppressed the leukemia burden of an RARA SE+ pAML patient-derived xenograft mouse model compared with a RARA SE- patient-derived xenograft. Our work shows that examining chromatin regulation can identify new, druggable dependencies in pAML and provides a rationale for a pediatric tamibarotene trial in children with RARA-high AML.
© 2021 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures

References
-
- Zwaan CM, Meshinchi S, Radich JP, et al. FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: prognostic significance and relation to cellular drug resistance. Blood. 2003;102(7):2387-2394. - PubMed
-
- Milligan DW, Wheatley K, Littlewood T, Craig JI, Burnett AK, Group NHOCS; NCRI Haematological Oncology Clinical Studies Group .Fludarabine and cytosine are less effective than standard ADE chemotherapy in high-risk acute myeloid leukemia, and addition of G-CSF and ATRA are not beneficial: results of the MRC AML-HR randomized trial. Blood. 2006;107(12):4614-4622. - PubMed
-
- Schlenk RF, Lübbert M, Benner A, et al. ; German-Austrian Acute Myeloid Leukemia Study Group . All-trans retinoic acid as adjunct to intensive treatment in younger adult patients with acute myeloid leukemia: results of the randomized AMLSG 07-04 study. Ann Hematol. 2016;95(12): 1931-1942. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

