High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape
- PMID: 34544114
- PMCID: PMC9241107
- DOI: 10.1038/s41586-021-04005-0
High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape
Abstract
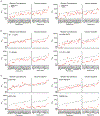
The number and variability of the neutralizing epitopes targeted by polyclonal antibodies in individuals who are SARS-CoV-2 convalescent and vaccinated are key determinants of neutralization breadth and the genetic barrier to viral escape1-4. Using HIV-1 pseudotypes and plasma selection experiments with vesicular stomatitis virus/SARS-CoV-2 chimaeras5, here we show that multiple neutralizing epitopes, within and outside the receptor-binding domain, are variably targeted by human polyclonal antibodies. Antibody targets coincide with spike sequences that are enriched for diversity in natural SARS-CoV-2 populations. By combining plasma-selected spike substitutions, we generated synthetic 'polymutant' spike protein pseudotypes that resisted polyclonal antibody neutralization to a similar degree as circulating variants of concern. By aggregating variant of concern-associated and antibody-selected spike substitutions into a single polymutant spike protein, we show that 20 naturally occurring mutations in the SARS-CoV-2 spike protein are sufficient to generate pseudotypes with near-complete resistance to the polyclonal neutralizing antibodies generated by individuals who are convalescent or recipients who received an mRNA vaccine. However, plasma from individuals who had been infected and subsequently received mRNA vaccination neutralized pseudotypes bearing this highly resistant SARS-CoV-2 polymutant spike, or diverse sarbecovirus spike proteins. Thus, optimally elicited human polyclonal antibodies against SARS-CoV-2 should be resilient to substantial future SARS-CoV-2 variation and may confer protection against potential future sarbecovirus pandemics.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures

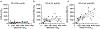









Comment in
-
COVID super-immunity: one of the pandemic's great puzzles.Nature. 2021 Oct;598(7881):393-394. doi: 10.1038/d41586-021-02795-x. Nature. 2021. PMID: 34650244 No abstract available.
References
Methods references
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

