SARS-CoV-2 Nsp5 Demonstrates Two Distinct Mechanisms Targeting RIG-I and MAVS To Evade the Innate Immune Response
- PMID: 34544279
- PMCID: PMC8546575
- DOI: 10.1128/mBio.02335-21
SARS-CoV-2 Nsp5 Demonstrates Two Distinct Mechanisms Targeting RIG-I and MAVS To Evade the Innate Immune Response
Abstract
Newly emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused a global pandemic with astonishing mortality and morbidity. The high replication and transmission of SARS-CoV-2 are remarkably distinct from those of previous closely related coronaviruses, and the underlying molecular mechanisms remain unclear. The innate immune defense is a physical barrier that restricts viral replication. We report here that the SARS-CoV-2 Nsp5 main protease targets RIG-I and mitochondrial antiviral signaling (MAVS) protein via two distinct mechanisms for inhibition. Specifically, Nsp5 cleaves off the 10 most-N-terminal amino acids from RIG-I and deprives it of the ability to activate MAVS, whereas Nsp5 promotes the ubiquitination and proteosome-mediated degradation of MAVS. As such, Nsp5 potently inhibits interferon (IFN) induction by double-stranded RNA (dsRNA) in an enzyme-dependent manner. A synthetic small-molecule inhibitor blunts the Nsp5-mediated destruction of cellular RIG-I and MAVS and processing of SARS-CoV-2 nonstructural proteins, thus restoring the innate immune response and impeding SARS-CoV-2 replication. This work offers new insight into the immune evasion strategy of SARS-CoV-2 and provides a potential antiviral agent to treat CoV disease 2019 (COVID-19) patients. IMPORTANCE The ongoing COVID-19 pandemic is caused by SARS-CoV-2, which is rapidly evolving with better transmissibility. Understanding the molecular basis of the SARS-CoV-2 interaction with host cells is of paramount significance, and development of antiviral agents provides new avenues to prevent and treat COVID-19 diseases. This study describes a molecular characterization of innate immune evasion mediated by the SARS-CoV-2 Nsp5 main protease and subsequent development of a small-molecule inhibitor.
Keywords: E3 ligase; MAVS; Nsp5; RIG-I; SARS-CoV-2; protease; small-molecule inhibitor.
Figures


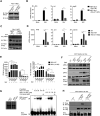



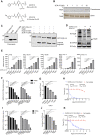
References
-
- Hui DS, I Azhar E, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh TD, Memish ZA, Drosten C, Zumla A, Petersen E. 2020. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91:264–266. doi:10.1016/j.ijid.2020.01.009. - DOI - PMC - PubMed
-
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, C4591001 Clinical Trial Group . 2020. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. doi:10.1056/NEJMoa2034577. - DOI - PMC - PubMed
-
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T. 2021. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 384:403–416. doi:10.1056/NEJMoa2035389. - DOI - PMC - PubMed
-
- Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, Bibi S, Briner C, Cicconi P, Collins AM, Colin-Jones R, Cutland CL, Darton TC, Dheda K, Duncan CJA, Emary KRW, Ewer KJ, Fairlie L, Faust SN, Feng S, Ferreira DM, Finn A, Goodman AL, Green CM, Green CA, Heath PT, Hill C, Hill H, Hirsch I, Hodgson SHC, Izu A, Jackson S, Jenkin D, Joe CCD, Kerridge S, Koen A, Kwatra G, Lazarus R, Lawrie AM, Lelliott A, Libri V, Lillie PJ, Mallory R, Mendes AVA, Milan EP, Minassian AM, et al. . 2021. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK Lancet 397:99–111. doi:10.1016/S0140-6736(20)32661-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

