IL13Rα2 Promotes Proliferation and Outgrowth of Breast Cancer Brain Metastases
- PMID: 34544797
- PMCID: PMC8595859
- DOI: 10.1158/1078-0432.CCR-21-0361
IL13Rα2 Promotes Proliferation and Outgrowth of Breast Cancer Brain Metastases
Abstract
Purpose: The survival of women with brain metastases (BM) from breast cancer remains very poor, with over 80% dying within a year of their diagnosis. Here, we define the function of IL13Rα2 in outgrowth of breast cancer brain metastases (BCBM) in vitro and in vivo, and postulate IL13Rα2 as a suitable therapeutic target for BM.
Experimental design: We performed IHC staining of IL13Rα2 in BCBM to define its prognostic value. Using inducible shRNAs in TNBC and HER2+ breast-brain metastatic models, we assessed IL13Rα2 function in vitro and in vivo. We performed RNAseq and functional studies to define the molecular mechanisms underlying IL13Rα2 function in BCBM.
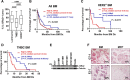
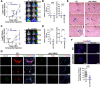
Results: High IL13Rα2 expression in BCBM predicted worse survival after BM diagnoses. IL13Rα2 was essential for cancer-cell survival, promoting proliferation while repressing invasion. IL13Rα2 KD resulted in FAK downregulation, repression of cell cycle and proliferation mediators, and upregulation of Ephrin B1 signaling. Ephrin-B1 (i) promoted invasion of BC cells in vitro, (ii) marked micrometastasis and invasive fronts in BCBM, and (iii) predicted shorter disease-free survival and BM-free survival (BMFS) in breast primary tumors known to metastasize to the brain. In experimental metastases models, which bypass early tumor invasion, downregulation of IL13Rα2 before or after tumor seeding and brain intravasation decreased BMs, suggesting that IL13Rα2 and the promotion of a proliferative phenotype is critical to BM progression.
Conclusions: Non-genomic phenotypic adaptations at metastatic sites are critical to BM progression and patients' prognosis. This study opens the road to use IL13Rα2 targeting as a therapeutic strategy for BM.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures




Similar articles
-
Targeting IL13Ralpha2 activates STAT6-TP63 pathway to suppress breast cancer lung metastasis.Breast Cancer Res. 2015 Jul 25;17(1):98. doi: 10.1186/s13058-015-0607-y. Breast Cancer Res. 2015. PMID: 26208975 Free PMC article.
-
An IL13Rα2 peptide exhibits therapeutic activity against metastatic colorectal cancer.Br J Cancer. 2018 Oct;119(8):940-949. doi: 10.1038/s41416-018-0259-7. Epub 2018 Oct 15. Br J Cancer. 2018. PMID: 30318506 Free PMC article.
-
Interleukin 13 receptor alpha 2 (IL13Rα2): Expression, signaling pathways and therapeutic applications in cancer.Biochim Biophys Acta Rev Cancer. 2022 Sep;1877(5):188802. doi: 10.1016/j.bbcan.2022.188802. Epub 2022 Sep 21. Biochim Biophys Acta Rev Cancer. 2022. PMID: 36152905 Review.
-
Glioma IL13Rα2 is associated with mesenchymal signature gene expression and poor patient prognosis.PLoS One. 2013 Oct 18;8(10):e77769. doi: 10.1371/journal.pone.0077769. eCollection 2013. PLoS One. 2013. PMID: 24204956 Free PMC article.
-
Treatment strategies for breast cancer brain metastases.Br J Cancer. 2021 Jan;124(1):142-155. doi: 10.1038/s41416-020-01175-y. Epub 2020 Nov 30. Br J Cancer. 2021. PMID: 33250512 Free PMC article. Review.
Cited by
-
An intraepithelial ILC1-like natural killer cell subset produces IL-13.Front Immunol. 2025 Mar 6;16:1521086. doi: 10.3389/fimmu.2025.1521086. eCollection 2025. Front Immunol. 2025. PMID: 40114916 Free PMC article.
-
Transcriptomics of Canine Inflammatory Mammary Cancer Treated with Empty Cowpea Mosaic Virus Implicates Neutrophils in Anti-Tumor Immunity.Int J Mol Sci. 2023 Sep 13;24(18):14034. doi: 10.3390/ijms241814034. Int J Mol Sci. 2023. PMID: 37762335 Free PMC article.
-
Targeting MTAP increases PARP inhibitor susceptibility in triple-negative breast cancer through a feed-forward loop.J Clin Invest. 2025 Jul 1;135(13):e188120. doi: 10.1172/JCI188120. eCollection 2025 Jul 1. J Clin Invest. 2025. PMID: 40590219 Free PMC article.
-
CREB5 promotes tumorigenicity and upregulates druggable cell surface modalities in basal-like breast cancer.NPJ Precis Oncol. 2025 Aug 6;9(1):276. doi: 10.1038/s41698-025-01051-z. NPJ Precis Oncol. 2025. PMID: 40770411 Free PMC article.
-
Protein Tyrosine Phosphatase 1B (PTP1B): A Comprehensive Review of Its Role in Pathogenesis of Human Diseases.Int J Mol Sci. 2024 Jun 27;25(13):7033. doi: 10.3390/ijms25137033. Int J Mol Sci. 2024. PMID: 39000142 Free PMC article. Review.
References
-
- Lee SS, Ahn J-H, Kim MK, Sym SJ, Gong G, Ahn SD, et al. Brain metastases in breast cancer: prognostic factors and management. Breast Cancer Res Treat 2008;111:523–30. - PubMed
-
- Palmieri D, Smith QR, Lockman PR, Bronder J, Gril B, Chambers AF, et al. Brain metastases of breast cancer. Breast Dis 2006;26:139–47. - PubMed
-
- Morris PG, Murphy CG, Mallam D, Accordino M, Patil S, Howard J, et al. Limited overall survival in patients with brain metastases from triple-negative breast cancer. Breast J 2012;18:345–50. - PubMed
-
- Barderas R, Bartolomé RA, Fernandez-Aceñero MJ, Torres S, Casal JI. High expression of IL-13 receptor α2 in colorectal cancer is associated with invasion, liver metastasis, and poor prognosis. Cancer Res 2012;72:2780–90. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

