Structural variants are a major source of gene expression differences in humans and often affect multiple nearby genes
- PMID: 34544830
- PMCID: PMC8647827
- DOI: 10.1101/gr.275488.121
Structural variants are a major source of gene expression differences in humans and often affect multiple nearby genes
Abstract
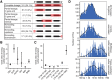
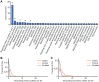
Structural variants (SVs) are an important source of human genome diversity, but their functional effects are poorly understood. We mapped 61,668 SVs in 613 individuals from the GTEx project and measured their effects on gene expression. We estimate that common SVs are causal at 2.66% of eQTLs, a 10.5-fold enrichment relative to their abundance in the genome. Duplications and deletions were the most impactful variant types, whereas the contribution of mobile element insertions was small (0.12% of eQTLs, 1.9-fold enriched). Multitissue analysis of eQTLs revealed that gene-altering SVs show more constitutive effects than other variant types, with 62.09% of coding SV-eQTLs active in all tissues with eQTL activity compared with 23.08% of coding SNV- and indel-eQTLs. Noncoding SVs, SNVs and indels show broadly similar patterns. We also identified 539 rare SVs associated with nearby gene expression outliers. Of these, 62.34% are noncoding SVs that affect gene expression but have modest enrichment at regulatory elements, showing that rare noncoding SVs are a major source of gene expression differences but remain difficult to predict from current annotations. Both common and rare SVs often affect the expression of multiple genes: SV-eQTLs affect an average of 1.82 nearby genes, whereas SNV- and indel-eQTLs affect an average of 1.09 genes, and 21.34% of rare expression-altering SVs show effects on two to nine different genes. We also observe significant effects on rare gene expression changes extending 1 Mb from the SV. This provides a mechanism by which individual SVs may have strong or pleiotropic effects on phenotypic variation.
© 2021 Scott et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
