Comprehensive molecular characterization of pediatric radiation-induced high-grade glioma
- PMID: 34545084
- PMCID: PMC8452624
- DOI: 10.1038/s41467-021-25709-x
Comprehensive molecular characterization of pediatric radiation-induced high-grade glioma
Abstract
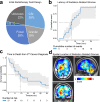
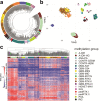
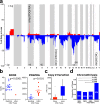
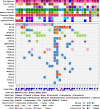
Radiation-induced high-grade gliomas (RIGs) are an incurable late complication of cranial radiation therapy. We performed DNA methylation profiling, RNA-seq, and DNA sequencing on 32 RIG tumors and an in vitro drug screen in two RIG cell lines. We report that based on DNA methylation, RIGs cluster primarily with the pediatric receptor tyrosine kinase I high-grade glioma subtype. Common copy-number alterations include Chromosome (Ch.) 1p loss/1q gain, and Ch. 13q and Ch. 14q loss; focal alterations include PDGFRA and CDK4 gain and CDKN2A and BCOR loss. Transcriptomically, RIGs comprise a stem-like subgroup with lesser mutation burden and Ch. 1p loss and a pro-inflammatory subgroup with greater mutation burden and depleted DNA repair gene expression. Chromothripsis in several RIG samples is associated with extrachromosomal circular DNA-mediated amplification of PDGFRA and CDK4. Drug screening suggests microtubule inhibitors/stabilizers, DNA-damaging agents, MEK inhibition, and, in the inflammatory subgroup, proteasome inhibitors, as potentially effective therapies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

