Clinical and molecular characterization of five Chinese patients with autosomal recessive osteopetrosis
- PMID: 34545712
- PMCID: PMC8606217
- DOI: 10.1002/mgg3.1815
Clinical and molecular characterization of five Chinese patients with autosomal recessive osteopetrosis
Abstract
Background: Osteopetrosis is characterized by increased bone density and bone marrow cavity stenosis due to a decrease in the number of osteoclasts or the dysfunction of their differentiation and absorption properties usually caused by biallelic variants of the TCIRG1 and CLCN7 genes.
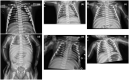
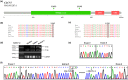
Methods: In this study, we describe five Chinese children who presented with anemia, thrombocytopenia, hepatosplenomegaly, repeated infections, and increased bone density. Whole-exome sequencing identified five compound heterozygous variants of the CLCN7 and TCIRG1 genes in these patients.
Results: Patient 1 had a novel variant c.1555C>T (p.L519F) and a previously reported pathogenic variant c.2299C>T (p.R767W) in CLCN7. Patient 2 harbored a novel missense variant (c.1025T>C; p.L342P) and a novel splicing variant (c.286-9G>A) in CLCN7. Patients 3A and 3B from one family displayed the same compound heterozygous TCIRG1 variant, including a novel frameshift variant (c.1370del; p.T457Tfs*71) and a novel splicing variant (c.1554+2T>C). In Patient 4, two novel variants were identified in the TCIRG1 gene: c.676G>T; p.E226* and c.1191del; p.P398Sfs*5. Patient 5 harbored two known pathogenic variants, c.909C>A (p.Y303*) and c.2008C>T (p.R670*), in TCIRG1. Analysis of the products obtained from the reverse transcription-polymerase chain reaction revealed that the c.286-9G>A variant in CLCN7 of patient 2 leads to intron 3 retention, resulting in the formation of a premature termination codon (p.E95Vfs*8). These five patients were eventually diagnosed with autosomal recessive osteopetrosis, and the three children with TCIRG1 variants received hematopoietic stem cell transplantation.
Conclusions: Our results expand the spectrum of variation of genes related to osteopetrosis and deepen the understanding of the relationship between the genotype and clinical characteristics of osteopetrosis.
Keywords: CLCN7; TCIRG1; autosomal recessive osteopetrosis; cDNA sequencing; novel variant.
© 2021 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures

Similar articles
-
Molecular Mechanisms of Craniofacial and Dental Abnormalities in Osteopetrosis.Int J Mol Sci. 2023 Jun 20;24(12):10412. doi: 10.3390/ijms241210412. Int J Mol Sci. 2023. PMID: 37373559 Free PMC article. Review.
-
Identification of TCIRG1 and CLCN7 gene mutations in a patient with autosomal recessive osteopetrosis.Mol Med Rep. 2014 Apr;9(4):1191-6. doi: 10.3892/mmr.2014.1955. Epub 2014 Feb 17. Mol Med Rep. 2014. PMID: 24535484
-
CLCN7 and TCIRG1 mutations differentially affect bone matrix mineralization in osteopetrotic individuals.J Bone Miner Res. 2014 Apr;29(4):982-91. doi: 10.1002/jbmr.2100. J Bone Miner Res. 2014. PMID: 24108692
-
Chloride channel ClCN7 mutations are responsible for severe recessive, dominant, and intermediate osteopetrosis.J Bone Miner Res. 2003 Oct;18(10):1740-7. doi: 10.1359/jbmr.2003.18.10.1740. J Bone Miner Res. 2003. PMID: 14584882
-
Rare gross deletion in T-cell immune regulator-1 gene in Iranian family with infantile malignant osteopetrosis.Saudi Med J. 2008 Oct;29(10):1494-6. Saudi Med J. 2008. PMID: 18946580 Review.
Cited by
-
Successful complete oral rehabilitation of a patient with osteopetrosis with extensive pre-treatments, bone grafts, dental implants and fixed bridges: a multidisciplinary case report.BMC Oral Health. 2023 Nov 28;23(1):940. doi: 10.1186/s12903-023-03707-3. BMC Oral Health. 2023. PMID: 38017429 Free PMC article.
-
Case report: Gene mutations and clinical characteristics of four patients with osteopetrosis.Front Pediatr. 2023 Mar 14;11:1096770. doi: 10.3389/fped.2023.1096770. eCollection 2023. Front Pediatr. 2023. PMID: 36999084 Free PMC article.
-
Molecular Mechanisms of Craniofacial and Dental Abnormalities in Osteopetrosis.Int J Mol Sci. 2023 Jun 20;24(12):10412. doi: 10.3390/ijms241210412. Int J Mol Sci. 2023. PMID: 37373559 Free PMC article. Review.
-
A novel compound heterozygous mutation of the CLCN7 gene is associated with autosomal recessive osteopetrosis.Front Pediatr. 2023 Apr 24;11:978879. doi: 10.3389/fped.2023.978879. eCollection 2023. Front Pediatr. 2023. PMID: 37168803 Free PMC article.
References
-
- Ajmal, M. , Mir, A. , Wahid, S. , Khor, C. C. , Foo, J. N. , Siddiqi, S. , Kauser, M. , Malik, S. A. , & Nasir, M. (2017). Identification and in silico characterization of a novel p.P208PfsX1 mutation in V‐ATPase a3 subunit associated with autosomal recessive osteopetrosis in a Pakistani family. BMC Medical Genetics, 18(1), 148. 10.1186/s12881-017-0506-4 - DOI - PMC - PubMed
-
- Bliznetz, E. A. , Tverskaya, S. M. , Zinchenko, R. A. , Abrukova, A. V. , Savaskina, E. N. , Nikulin, M. V. , Kirillov, A. G. , Ginter, E. K. , & Polyakov, A. V. (2009). Genetic analysis of autosomal recessive osteopetrosis in Chuvashiya: The unique splice site mutation in TCIRG1 gene spread by the founder effect. European Journal of Human Genetics, 17(5), 664–672. 10.1038/ejhg.2008.234 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

