Uncertainty in denoising of MRSI using low-rank methods
- PMID: 34545962
- PMCID: PMC7612041
- DOI: 10.1002/mrm.29018
Uncertainty in denoising of MRSI using low-rank methods
Abstract
Purpose: Low-rank denoising of MRSI data results in an apparent increase in spectral SNR. However, it is not clear if this translates to a lower uncertainty in metabolite concentrations after spectroscopic fitting. Estimation of the true uncertainty after denoising is desirable for downstream analysis in spectroscopy. In this work, the uncertainty reduction from low-rank denoising methods based on spatiotemporal separability and linear predictability in MRSI are assessed. A new method for estimating metabolite concentration uncertainty after denoising is proposed. Automatic rank threshold selection methods are also assessed in simulated low SNR regimes.
Methods: Assessment of denoising methods is conducted using Monte Carlo simulation of proton MRSI data and by reproducibility of repeated in vivo acquisitions in 5 subjects.
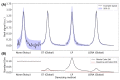
Results: In simulated and in vivo data, spatiotemporal based denoising is shown to reduce the concentration uncertainty, but linear prediction denoising increases uncertainty. Uncertainty estimates provided by fitting algorithms after denoising consistently underestimate actual metabolite uncertainty. However, the proposed uncertainty estimation, based on an analytical expression for entry-wise variance after denoising, is more accurate. It is also shown automated rank threshold selection using Marchenko-Pastur distribution can bias the data in low SNR conditions. An alternative soft-thresholding function is proposed.
Conclusion: Low-rank denoising methods based on spatiotemporal separability do reduce uncertainty in MRS(I) data. However, thorough assessment is needed as assessment by SNR measured from residual baseline noise is insufficient given the presence of non-uniform variance. It is also important to select the right rank thresholding method in low SNR cases.
Keywords: MRS; MRSI; denoising; low rank; spectroscopy.
© 2021 The Authors. Magnetic Resonance in Medicine published by Wiley Periodicals LLC on behalf of International Society for Magnetic Resonance in Medicine.
Figures








References
-
- Vizioli L, Moeller S, Dowdle L, et al. A paradigm change in functional brain mapping: suppressing the thermal noise in fMRI. bioRxiv. 2020:2020.11.04.368357
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

