A surrogate cell-based SARS-CoV-2 spike blocking assay
- PMID: 34547822
- PMCID: PMC8646767
- DOI: 10.1002/eji.202149302
A surrogate cell-based SARS-CoV-2 spike blocking assay
Abstract
To monitor infection by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and successful vaccination against coronavirus disease 2019 (COVID-19), the kinetics of neutralizing or blocking anti-SARS-CoV-2 antibody titers need to be assessed. Here, we report the development of a quick and inexpensive surrogate SARS-CoV-2 blocking assay (SUBA) using immobilized recombinant human angiotensin-converting enzyme 2 (hACE2) and human cells expressing the native form of surface SARS-CoV-2 spike protein. Spike protein-expressing cells bound to hACE2 in the absence or presence of blocking antibodies were quantified by measuring the optical density of cell-associated crystal violet in a spectrophotometer. The advantages are that SUBA is a fast and inexpensive assay, which does not require biosafety level 2- or 3-approved laboratories. Most importantly, SUBA detects blocking antibodies against the native trimeric cell-bound SARS-CoV-2 spike protein and can be rapidly adjusted to quickly pre-screen already approved therapeutic antibodies or sera from vaccinated individuals for their ACE2 blocking activities against any emerging SARS-CoV-2 variants.
Keywords: COVID-19; SARS-CoV-2; Spike protein; Surrogate blocking assay; hACE2.
© 2021 The Authors. European Journal of Immunology published by Wiley-VCH GmbH.
Conflict of interest statement
DM and HMJ have filed a patent application for SUBA. All other authors have no commercial or financial conflict of interests.
Figures
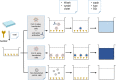
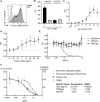


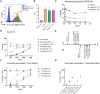
References
-
- To, K. K.‐W. , Tsang, O. T.‐Y. , Leung, W.‐S. , Tam, A. R. , Wu, T.‐C. , Lung, D. C. , Yip, C. C.‐Y. et al., Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect. Dis. 2020. 20(5): 565–574. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

