A single-cell tumor immune atlas for precision oncology
- PMID: 34548323
- PMCID: PMC8494216
- DOI: 10.1101/gr.273300.120
A single-cell tumor immune atlas for precision oncology
Abstract
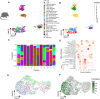
The tumor immune microenvironment is a main contributor to cancer progression and a promising therapeutic target for oncology. However, immune microenvironments vary profoundly between patients, and biomarkers for prognosis and treatment response lack precision. A comprehensive compendium of tumor immune cells is required to pinpoint predictive cellular states and their spatial localization. We generated a single-cell tumor immune atlas, jointly analyzing published data sets of >500,000 cells from 217 patients and 13 cancer types, providing the basis for a patient stratification based on immune cell compositions. Projecting immune cells from external tumors onto the atlas facilitated an automated cell annotation system. To enable in situ mapping of immune populations for digital pathology, we applied SPOTlight, combining single-cell and spatial transcriptomics data and identifying colocalization patterns of immune, stromal, and cancer cells in tumor sections. We expect the tumor immune cell atlas, together with our versatile toolbox for precision oncology, to advance currently applied stratification approaches for prognosis and immunotherapy.
© 2021 Nieto et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
