The human liver microenvironment shapes the homing and function of CD4+ T-cell populations
- PMID: 34548339
- PMCID: PMC9185819
- DOI: 10.1136/gutjnl-2020-323771
The human liver microenvironment shapes the homing and function of CD4+ T-cell populations
Abstract
Objective: Tissue-resident memory T cells (TRM) are vital immune sentinels that provide protective immunity. While hepatic CD8+ TRM have been well described, little is known about the location, phenotype and function of CD4+ TRM.
Design: We used multiparametric flow cytometry, histological assessment and novel human tissue coculture systems to interrogate the ex vivo phenotype, function and generation of the intrahepatic CD4+ T-cell compartment. We also used leukocytes isolated from human leukocyte antigen (HLA)-disparate liver allografts to assess long-term retention.
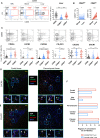
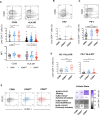
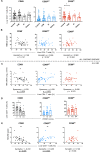
Results: Hepatic CD4+ T cells were delineated into three distinct populations based on CD69 expression: CD69-, CD69INT and CD69HI. CD69HICD4+ cells were identified as tissue-resident CD4+ T cells on the basis of their exclusion from the circulation, phenotypical profile (CXCR6+CD49a+S1PR1-PD-1+) and long-term persistence within the pool of donor-derived leukcoocytes in HLA-disparate liver allografts. CD69HICD4+ T cells produced robust type 1 polyfunctional cytokine responses on stimulation. Conversely, CD69INTCD4+ T cells represented a more heterogenous population containing cells with a more activated phenotype, a distinct chemokine receptor profile (CX3CR1+CXCR3+CXCR1+) and a bias towards interleukin-4 production. While CD69INTCD4+ T cells could be found in the circulation and lymph nodes, these cells also formed part of the long-term resident pool, persisting in HLA-mismatched allografts. Notably, frequencies of CD69INTCD4+ T cells correlated with necroinflammatory scores in chronic hepatitis B infection. Finally, we demonstrated that interaction with hepatic epithelia was sufficient to generate CD69INTCD4+ T cells, while additional signals from the liver microenvironment were required to generate liver-resident CD69HICD4+ T cells.
Conclusions: High and intermediate CD69 expressions mark human hepatic CD4+ TRM and a novel functionally distinct recirculating population, respectively, both shaped by the liver microenvironment to achieve diverse immunosurveillance.
Keywords: T lymphocytes; cellular immunity; hepatitis B; immunology; liver immunology.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: BW collaborated with and received funding from Bioniz. LJP sat on advisory boards/provided consultancy for Gilead Sciences and SQZ Biotech. KA was funded by a studentship with Dr Falk. MKM received research funding from Gilead Sciences, Hoffmann La Roche and Immunocore. MKM sat on advisory boards/provided consultancy for Gilead, Hoffmann La Roche, Immunocore, VIR, Galapagos NV, GSK, Abbvie and Freeline. ZS collaborated with Bioniz and AstraZeneca and consulted for Boehringer Ingelheim. All other authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
