Complete genome sequencing and molecular characterization of SARS-COV-2 from COVID-19 cases in Alborz province in Iran
- PMID: 34549097
- PMCID: PMC8447724
- DOI: 10.1016/j.heliyon.2021.e08027
Complete genome sequencing and molecular characterization of SARS-COV-2 from COVID-19 cases in Alborz province in Iran
Abstract
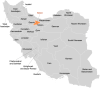
Iran was among countries which was hard hit at the early stage of the coronavirus disease 2019 (COVID-19) pandemic and dealt with the second wave of the pandemic in May and June 2020; however, there are a very limited number of complete genome sequences of acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from Iran. In this study, complete genome sequences of the virus in the samples obtained from three patients in Alborz province in May and June 2020 were generated and analyzed using bioinformatic methods. The sequenced genomes were positioned in a cluster with B.4 lineage along with the sequences from other countries namely, United Arab Emirates and Oman. There were seven single nucleotide variations (SNVs) in common in all samples and only one of the sequenced genomes showed the D614G amino acid substitution. Three SNVs, 1397 G > A, 28688T > C, 29742 G > T, which had already been reported in February, were found with high frequency in all the sequenced genomes in this study, implying that viral diversity reflected in the early stages of viral transmission in Iran were established in the second wave. Considering the importance of molecular epidemiology in response to ongoing pandemic, there is an urgent need for more complete genome sequencing and comprehensive analyses to gain insight into the transmission, adaptation and evolution of the virus in Iran.
Keywords: COVID-19; Clade; Lineage; Phylogenetic; SARS-CoV-2; Single nucleotide variation.
© 2021 The Authors.
Figures
Similar articles
-
Molecular epidemiology of SARS-CoV-2 isolated from COVID-19 family clusters.BMC Med Genomics. 2021 Jun 1;14(1):144. doi: 10.1186/s12920-021-00990-3. BMC Med Genomics. 2021. PMID: 34074255 Free PMC article.
-
Whole-genome sequencing of SARS-CoV-2 reveals the detection of G614 variant in Pakistan.PLoS One. 2021 Mar 23;16(3):e0248371. doi: 10.1371/journal.pone.0248371. eCollection 2021. PLoS One. 2021. PMID: 33755704 Free PMC article.
-
Mutations and Phylogenetic Analyses of SARS-CoV-2 Among Imported COVID-19 From Abroad in Nanjing, China.Front Microbiol. 2022 Mar 17;13:851323. doi: 10.3389/fmicb.2022.851323. eCollection 2022. Front Microbiol. 2022. PMID: 35369437 Free PMC article.
-
Characterization and phylogenetic analysis of Iranian SARS-CoV-2 genomes: A phylogenomic study.Health Sci Rep. 2023 Jan 16;6(1):e1052. doi: 10.1002/hsr2.1052. eCollection 2023 Jan. Health Sci Rep. 2023. PMID: 36686884 Free PMC article.
-
Genomic Evolution and Variation of SARS-CoV-2 in the Early Phase of COVID-19 Pandemic in Guangdong Province, China.Curr Med Sci. 2021 Apr;41(2):228-235. doi: 10.1007/s11596-021-2340-3. Epub 2021 Apr 20. Curr Med Sci. 2021. PMID: 33877539 Free PMC article.
Cited by
-
Improvement of the inactivated SARS-CoV-2 vaccine potency through formulation in alum/naloxone adjuvant; Robust T cell and anti-RBD IgG responses.Iran J Basic Med Sci. 2022 May;25(5):554-561. doi: 10.22038/IJBMS.2022.63527.14015. Iran J Basic Med Sci. 2022. PMID: 35911642 Free PMC article.
References
-
- Ghadir M.R., Ebrazeh A., Khodadadi J. The COVID-19 outbreak in Iran; the first patient with a definite diagnosis. Arch. Iran. Med. 2020;23:503–504. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

