The Vancomycin Resistance-Associated Regulatory System VraSR Modulates Biofilm Formation of Staphylococcus epidermidis in an ica-Dependent Manner
- PMID: 34550006
- PMCID: PMC8550092
- DOI: 10.1128/mSphere.00641-21
The Vancomycin Resistance-Associated Regulatory System VraSR Modulates Biofilm Formation of Staphylococcus epidermidis in an ica-Dependent Manner
Abstract
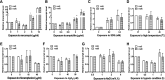
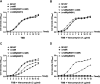
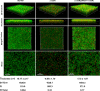
The two-component system VraSR responds to the cell wall-active antibiotic stress in Staphylococcus epidermidis. To study its regulatory function in biofilm formation, a vraSR deletion mutant (ΔvraSR) was constructed using S. epidermidis strain 1457 (SE1457) as the parent strain. Compared to SE1457, the ΔvraSR mutant showed impaired biofilm formation both in vitro and in vivo with a higher ratio of dead cells within the biofilm. Consistently, the ΔvraSR mutant produced much less polysaccharide intercellular adhesin (PIA). The ΔvraSR mutant also showed increased susceptibility to the cell wall inhibitor and SDS, and its cell wall observed under a transmission electron microscope (TEM) appeared to be thinner and interrupted, which is in accordance with higher susceptibility to the stress. Complementation of vraSR in the ΔvraSR mutant restored the biofilm formation and the cell wall thickness to wild-type levels. Transcriptome sequencing (RNA-Seq) showed that the vraSR deletion affected the transcription levels of 73 genes, including genes involved in biofilm formation, bacterial programmed cell death (CidA-LrgAB system), glycolysis/gluconeogenesis, the pentose phosphate pathway (PPP), and the tricarboxylic acid (TCA) cycle, etc. The results of RNA-Seq were confirmed by quantitative real-time reverse transcription-PCR (qRT-PCR). In the ΔvraSR mutant, the expression of icaA and lrgAB was downregulated and the expression of icaR and cidA was upregulated, in comparison to that of SE1457. The transcriptional levels of antibiotic-resistant genes (pbp2, serp1412, murAA, etc.) had no significant changes. An electrophoretic mobility shift assay further revealed that phosphorylated VraR bound to the promoter regions of the ica operon, as well as its own promoter region. This study demonstrates that in S. epidermidis, VraSR is an autoregulator and directly regulates biofilm formation in an ica-dependent manner. Upon cell wall stress, it indirectly regulates cell death and drug resistance in association with alterations to multiple metabolism pathways. IMPORTANCE S. epidermidis is a leading cause of hospital-acquired catheter-related infections, and its pathogenicity depends mostly on its ability to form biofilms on implants. The biofilm formation is a complex procedure that involves multiple regulating factors. Here, we show that a vancomycin resistance-associated two-component regulatory system, VraSR, plays an important role in modulating S. epidermidis biofilm formation and tolerance to stress. We demonstrate that S. epidermidis VraSR is an autoregulated system that selectively responds to stress targeting cell wall synthesis. Besides, phosphorylated VraR can bind to the promoter region of the ica operon and directly regulates polysaccharide intercellular adhesin production and biofilm formation in S. epidermidis. Furthermore, VraSR may indirectly modulate bacterial cell death and extracellular DNA (eDNA) release in biofilms through the CidA-LrgAB system. This work provides a new molecular insight into the mechanisms of VraSR-mediated modulation of the biofilm formation and cell death of S. epidermidis.
Keywords: Staphylococcus epidermidis; VraSR; biofilm formation; cell death; two-component regulatory system.
Figures






References
-
- O'Connor AM, McManus BA, Kinnevey PM, Brennan GI, Fleming TE, Cashin PJ, O'Sullivan M, Polyzois I, Coleman DC. 2018. Significant enrichment and diversity of the staphylococcal arginine catabolic mobile element ACME in Staphylococcus epidermidis isolates from subgingival peri-implantitis sites and periodontal pockets. Front Microbiol 9:1558. doi: 10.3389/fmicb.2018.01558. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

