DNA damage-induced degradation of Sp1 promotes cellular senescence
- PMID: 34550526
- PMCID: PMC9135943
- DOI: 10.1007/s11357-021-00456-5
DNA damage-induced degradation of Sp1 promotes cellular senescence
Abstract
Persistent DNA damage (genotoxic stress) triggers signaling cascades that drive cells into apoptosis or senescence to avoid replicating a damaged genome. Sp1 has been found to play a role in double strand break (DSB) repair, and a link between Sp1 and aging has also been established, where Sp1 protein, but not RNA, levels decrease with age. Interestingly, inhibition ATM reverses the age-related degradation of Sp1, suggesting that DNA damage signaling is involved in senescence-related degradation of Sp1. Proteasomal degradation of Sp1 in senescent cells is mediated via sumoylation, where sumoylation of Sp1 on lysine 16 is increased in senescent cells. Taking into consideration our previous findings that Sp1 is phosphorylated by ATM in response to DNA damage and that proteasomal degradation of Sp1 at DSBs is also mediated by its sumoylation and subsequent interaction with RNF4, we investigated the potential contribution of Sp1's role as a DSB repair factor in mediating cellular senescence. We report here that Sp1 expression is decreased with a concomitant increase in senescence markers in response to DNA damage. Mutation of Sp1 at serine 101 to create an ATM phospho-null mutant, or mutation of lysine 16 to create a sumo-null mutant, prevents the sumoylation and subsequent proteasomal degradation of Sp1 and results in a decrease in senescence. Conversely, depletion of Sp1 or mutation of Sp1 to create an ATM phosphomimetic results in premature degradation of Sp1 and an increase in senescence markers. These data link a loss of genomic stability with senescence through the action of a DNA damage repair factor.
Keywords: DNA damage; SUMOylation; Senescence; Sp1; Ubiquitylation.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

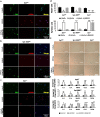

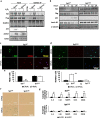

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

