High-throughput single-cell quantification of hundreds of proteins using conventional flow cytometry and machine learning
- PMID: 34550730
- PMCID: PMC8457665
- DOI: 10.1126/sciadv.abg0505
High-throughput single-cell quantification of hundreds of proteins using conventional flow cytometry and machine learning
Abstract
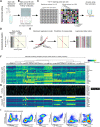
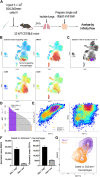
Modern immunologic research increasingly requires high-dimensional analyses to understand the complex milieu of cell types that comprise the tissue microenvironments of disease. To achieve this, we developed Infinity Flow combining hundreds of overlapping flow cytometry panels using machine learning to enable the simultaneous analysis of the coexpression patterns of hundreds of surface-expressed proteins across millions of individual cells. In this study, we demonstrate that this approach allows the comprehensive analysis of the cellular constituency of the steady-state murine lung and the identification of previously unknown cellular heterogeneity in the lungs of melanoma metastasis–bearing mice. We show that by using supervised machine learning, Infinity Flow enhances the accuracy and depth of clustering or dimensionality reduction algorithms. Infinity Flow is a highly scalable, low-cost, and accessible solution to single-cell proteomics in complex tissues.
Figures



References
-
- Engel P., Boumsell L., Balderas R., Bensussan A., Gattei V., Horejsi V., Jin B. Q., Malavasi F., Mortari F., Schwartz-Albiez R., Stockinger H., van Zelm M. C., Zola H., Clark G., Cd nomenclature 2015: Human leukocyte differentiation antigen workshops as a driving force in immunology. J. Immunol. 195, 4555–4563 (2015). - PubMed
-
- Pedreira C. E., Costa E. S., Barrena S., Lecrevisse Q., Almeida J., van Dongen J. J. M., Orfao A.; on behalf of EuroFlow Consortium , Generation of flow cytometry data files with a potentially infinite number of dimensions. Cytometry A 73A, 834–846 (2008). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

