Exogene: A performant workflow for detecting viral integrations from paired-end next-generation sequencing data
- PMID: 34550971
- PMCID: PMC8457494
- DOI: 10.1371/journal.pone.0250915
Exogene: A performant workflow for detecting viral integrations from paired-end next-generation sequencing data
Abstract
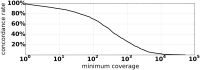
The integration of viruses into the human genome is known to be associated with tumorigenesis in many cancers, but the accurate detection of integration breakpoints from short read sequencing data is made difficult by human-viral homologies, viral genome heterogeneity, coverage limitations, and other factors. To address this, we present Exogene, a sensitive and efficient workflow for detecting viral integrations from paired-end next generation sequencing data. Exogene's read filtering and breakpoint detection strategies yield integration coordinates that are highly concordant with long read validation. We demonstrate this concordance across 6 TCGA Hepatocellular carcinoma (HCC) tumor samples, identifying integrations of hepatitis B virus that are also supported by long reads. Additionally, we applied Exogene to targeted capture data from 426 previously studied HCC samples, achieving 98.9% concordance with existing methods and identifying 238 high-confidence integrations that were not previously reported. Exogene is applicable to multiple types of paired-end sequence data, including genome, exome, RNA-Seq and targeted capture.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Henle G, Henle W, Clifford P, Diehl V, Kafuko GW, Kirya BG, et al.. Antibodies to Epstein-Barr virus in Burkitt’s lymphoma and control groups. Journal of the National Cancer Institute. 1969;43(5):1147–1157. - PubMed
-
- Nonoyama M, Kawai Y, Pagano J. Detection of Epstein-Barr virus DNA in human tumors. Bibliotheca Haematologica. 1975;40:577–583. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

