Ferritin heavy chain (FTH1) exerts significant antigrowth effects in breast cancer cells by inhibiting the expression of c-MYC
- PMID: 34551213
- PMCID: PMC8564339
- DOI: 10.1002/2211-5463.13303
Ferritin heavy chain (FTH1) exerts significant antigrowth effects in breast cancer cells by inhibiting the expression of c-MYC
Abstract
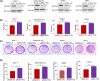
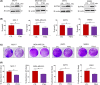
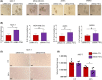
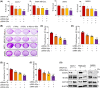
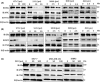
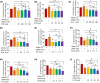
Overexpression of ferritin heavy chain (FTH1) often associates with good prognosis in breast cancer (BCa), particularly in the triple-negative subtype (triple-negative breast cancer). However, the mechanism by which FTH1 exerts its possible tumor suppressor effects in BCa is not known. Here, we examined the bearing of FTH1 silencing or overexpression on several aspects of BCa cell growth in vitro. FTH1 silencing promoted cell growth and mammosphere formation, increased c-MYC expression, and reduced cell sensitivity to chemotherapy. In contrast, FTH1 overexpression inhibited cell growth, decreased c-MYC expression, and sensitized cancer cells to chemotherapy; silencing of c-MYC recapitulated the effects of FTH1 overexpression. These findings show for the first time that FTH1 suppresses tumor growth by inhibiting the expression of key oncogenes, such as c-MYC.
Keywords: FTH1; G9a; breast cancer; c-MYC; iron metabolism.
© 2021 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict interest.
Figures


References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. - PubMed
-
- Harbeck N, Penault‐Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J and Cardoso F (2019) Breast cancer. Nat Rev Dis Primers 5, 66. - PubMed
-
- Harrison PM and Arosio P (1996) The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275, 161–203. - PubMed
-
- Lawson DM, Artymiuk PJ, Yewdall SJ, Smith JM, Livingstone JC, Treffry A, Luzzago A, Levi S, Arosio P and Cesareni G (1991) Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature 349, 541–544. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

