MYC Levels Regulate Metastatic Heterogeneity in Pancreatic Adenocarcinoma
- PMID: 34551968
- PMCID: PMC8831468
- DOI: 10.1158/2159-8290.CD-20-1826
MYC Levels Regulate Metastatic Heterogeneity in Pancreatic Adenocarcinoma
Abstract
The degree of metastatic disease varies widely among patients with cancer and affects clinical outcomes. However, the biological and functional differences that drive the extent of metastasis are poorly understood. We analyzed primary tumors and paired metastases using a multifluorescent lineage-labeled mouse model of pancreatic ductal adenocarcinoma (PDAC)-a tumor type in which most patients present with metastases. Genomic and transcriptomic analysis revealed an association between metastatic burden and gene amplification or transcriptional upregulation of MYC and its downstream targets. Functional experiments showed that MYC promotes metastasis by recruiting tumor-associated macrophages, leading to greater bloodstream intravasation. Consistent with these findings, metastatic progression in human PDAC was associated with activation of MYC signaling pathways and enrichment for MYC amplifications specifically in metastatic patients. Collectively, these results implicate MYC activity as a major determinant of metastatic burden in advanced PDAC. SIGNIFICANCE: Here, we investigate metastatic variation seen clinically in patients with PDAC and murine PDAC tumors and identify MYC as a major driver of this heterogeneity.This article is highlighted in the In This Issue feature, p. 275.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures
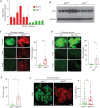
![Figure 1. Advanced pancreatic tumors exhibit intertumoral differences in their propensity for metastasis. A, CT imaging of human PDAC liver metastasis demonstrating heterogeneity in metastatic burden in stage IV disease. Arrowheads indicate solitary metastasis in the top image and selected metastases in the bottom. B, Density plot and histogram showing the distribution of total (liver and lung) metastases enumerated from CT scans of human stage IV PDAC at the time of diagnosis (n = 55). Values above each histogram bar represent the number of patients in each group. The vertical dotted line (red) represents the cutoff between MetLow tumors [≤10 metastases (mets)] and MetHigh tumors (>10 mets) determined by k-means clustering. C, Quantification of tumor area (based on tumor dimensions from largest cross-sectional plane on imaging) comparing MetLow and MetHigh cases from the cohort in B. D, Overall survival analysis of the cohort in B. E, Top, schematic view of the KPCXY model, showing multiple primary tumors distinguishable by color arising in the pancreas with matched metastases in the liver. Bottom, representative fluorescent stereomicroscopic images showing a YFP+ tumor adjoining a CFP+ tumor in the pancreas (left) and liver metastases derived from the CFP+ tumor in the same animal (right). F, Density plot and histogram showing the distribution of total (liver and lung) metastases enumerated at autopsy of KPCXY mice. Values above each histogram bar represent the number of tumors giving rise to the indicated number of metastases, based on color (n = 85 tumors from 30 KPCXY mice). The vertical dotted line (red) represents the cutoff between MetLow tumors (≤10 mets, n = 58) and MetHigh tumors (>10 mets, n = 28) determined by k-means clustering. G, Quantification of tumor area comparing MetLow and MetHigh tumors from the cohort in F. Statistical analysis by Student unpaired t test with P values indicated (ns, not significant). Box-and-whisker plots in C and G indicate mean and interquartile range. Scale bar for E, 1 mm.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8a7a/9651160/200760200a9c/542fig1.gif)

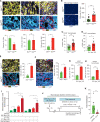
![Figure 3. The MetHigh phenotype is associated with focal, high-amplitude Myc amplifications and elevated expression. A, Schematic representation of focal amplifications identified in profiled primary tumors. Vertical gray line denotes the location of amplicon and likely driver gene. B, Zoomed-in schematic representation of three identified Myc amplicons in MetHigh tumors illustrating the focal and high-amplitude nature of the event (left). Each event (amplicon) is illustrated by a different colored segment line. The shared amplified region between the different amplicons is denoted by the chromosomal cytoband top of panel and illustrated in a UCSC Genome Browser view (right) with RefSeq genes, including Myc, illustrated. C, Box-and-whisker plot showing Myc mRNA levels in MetHigh tumors (n = 7) and paired metastases (n = 34) compared with MetLow tumors (n = 13). FPKM, fragments per kilobase of exon per million. D, Volcano plot illustrating genes meeting cutoffs for differential expression [log fold change (logFC) >1, Padj. < 0.05] between MetHigh and MetLow tumors (n = 20 tumors used in the comparison). Genes upregulated in MetHigh tumors are highlighted in green, and genes upregulated in MetLow tumors are highlighted in red. E, Top 10 hallmark gene sets identified as enriched in MetHigh tumors compared with MetLow tumors using all differentially expressed genes (DEG; Padj. < 0.05). F, Top five transcription factor (TF) binding sites enriched in DEGs in MetHigh tumors compared with MetLow tumors (Padj. < 0.05) identified by Metacore prediction software. G, Heat map showing unsupervised clustering of DEGs (logFC >1, Padj. < 0.05) between MetHigh and MetLow tumors (n = 20) and their association with PDAC transcriptional subtypes previously reported by Collisson and colleagues (42), Moffitt and colleagues (15), and Bailey and colleagues (16). ADEX, aberrantly differentiated endocrine exocrine; QM-PDA, quasi-mesenchymal-pancreatic ductal adenocarcinoma. H, Kaplan–Meier analysis showing overall survival of patients with PDAC in the TCGA cohort stratified into those with a MetHigh signature (red line) versus those with a MetLow signature (green line). Signature based on DEGs with absolute logFC >0.58 and Padj. < 0.05 (736 up- and 1,036 downregulated genes). Statistical analysis in C was performed by Wilcoxon test (*, P = 3.9 × 10−4; **, P = 5.3 × 10−5). Box and whiskers represent median mRNA expression and interquartile range. Statistical analysis in H was performed by log-rank test.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8a7a/9651160/5b7293a6054e/542fig3.gif)
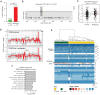
![Figure 6. MYC acts through CXCL3 and MIF to promote macrophage recruitment and metastasis. A, Expression of selected cytokines/chemokines in human PDAC. Samples from the COMPASS cohort (enriched for tumor cells by laser capture microdissection) were stratified into MYC-high and MYC-low groups based on RNA-seq (n = 373) and assessed for the expression of five chemokines/cytokines identified as significantly upregulated in MetHigh versus MetLow tumors (Supplementary Fig. S8A). FPKM, fragments per kilobase of exon per million. B, Relative expression of Mif and Cxcl3 in control or Myc knockdown [short hairpin RNA (shRNA)] MetHigh cell line 850_MetHigh_4. Data are representative of two independent Myc shRNAs (n = 3 biological replicates). C, Bar graph showing fold increase in Cxcl3 and Mif mRNA levels comparing Myc_OE to EV control cell lines. Data are representative of two independent cell lines (n = 3 biological replicates). D, Quantification of total F4/80+ tumor-infiltrating macrophages by immunoflourescence in cell lines that were stably transduced with either a Cxcl3 or Mif overexpression construct (Cxcl3_OE and Mif_OE, respectively) or empty vector (EV), with n = 4 tumors examined from each group with four to five random fields of view analyzed. E, Quantification of total metastases (liver and lung) following orthotopic transplantation of EV, Cxcl3_OE, or Mif_OE orthotopic tumors from D. Data were pooled from two independent MetLow lines transduced with the Cxcl3_OE, Mif_OE, or EV construct transplanted into five NOD.SCID mice (for each cell line). Each dot represents an independent animal. F, Quantification of macrophages that migrated across a transwell filter following coculture with 832 Myc_OE tumor cells treated with either a Cxcr2 inhibitor (AZD5069) or a MIF inhibitor (ISO-1). Data are representative of two independent experiments, including three replicates with four to five 20× images taken per transwell. G, Schematic outline of the CXCR2 and MIF inhibitor experiment. Mice were orthotopically implanted with 832 Myc_OE cells and after 10 days were treated with a CXCR2 inhibitor (AZD5069), MIF inhibitor (ISO-1), combination (AZD5069 + ISO-1), or vehicle. Metastases and macrophages were quantified 14 days later. H and I, Quantification of F4/80+ (H) and CD206 (I) macrophages in orthtotopic tumors following the CXCR2 and MIF strategy outlined in G (n = 4 tumors per group; four to five random fields of view analyzed; each dot represents an independent animal). J, Quantification of total metastases (liver and lung) following the CXCR2 and MIF strategy outlined in G (n = 4 control mice, n = 4 AZD5069 mice, n = 4 ISO-1 mice, and n = 4 AZD5069 + ISO-1 mice; each dot represents an independent animal). Statistical analysis by Student t test with significance indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.007; ****, P < 0.0005; *****, P < 0.0001; ns, not significant). Error bars indicate SEM.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8a7a/9651160/54885def35f2/542fig6.gif)
Comment in
- doi: 10.1158/2159-8290.CD-12-2-ITI
References
-
- McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

