Paths and timings of the peopling of Polynesia inferred from genomic networks
- PMID: 34552258
- PMCID: PMC9710236
- DOI: 10.1038/s41586-021-03902-8
Paths and timings of the peopling of Polynesia inferred from genomic networks
Abstract
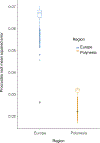
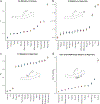
Polynesia was settled in a series of extraordinary voyages across an ocean spanning one third of the Earth1, but the sequences of islands settled remain unknown and their timings disputed. Currently, several centuries separate the dates suggested by different archaeological surveys2-4. Here, using genome-wide data from merely 430 modern individuals from 21 key Pacific island populations and novel ancestry-specific computational analyses, we unravel the detailed genetic history of this vast, dispersed island network. Our reconstruction of the branching Polynesian migration sequence reveals a serial founder expansion, characterized by directional loss of variants, that originated in Samoa and spread first through the Cook Islands (Rarotonga), then to the Society (Tōtaiete mā) Islands (11th century), the western Austral (Tuha'a Pae) Islands and Tuāmotu Archipelago (12th century), and finally to the widely separated, but genetically connected, megalithic statue-building cultures of the Marquesas (Te Henua 'Enana) Islands in the north, Raivavae in the south, and Easter Island (Rapa Nui), the easternmost of the Polynesian islands, settled in approximately AD 1200 via Mangareva.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures


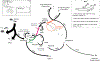

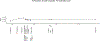

Comment in
-
Modern Polynesian genomes offer clues to early eastward migrations.Nature. 2021 Sep;597(7877):477-478. doi: 10.1038/d41586-021-01719-z. Nature. 2021. PMID: 34552247 No abstract available.
References
-
- Low S. Hawaiki Rising: Hōkūle’a, Nainoa Thompson, and the Hawaiian Renaissance. (University of Hawaii Press, 2019).
-
- Kirch PV On the Road of the Winds. (University of California Press, 2017).
-
- Schmid MME et al. How 14C dates on wood charcoal increase precision when dating colonization: The examples of Iceland and Polynesia. Quaternary Geochronology 48, 64–71 (2018).
-
- Kahōʻāliʻi Keauokalani K. Kepelino’s Traditions of Hawaii. Bernice P. Bishop Museum Bulletin; 206 (1932).

