Quantitative magnetic resonance imaging and tumor forecasting of breast cancer patients in the community setting
- PMID: 34552262
- PMCID: PMC9753909
- DOI: 10.1038/s41596-021-00617-y
Quantitative magnetic resonance imaging and tumor forecasting of breast cancer patients in the community setting
Abstract
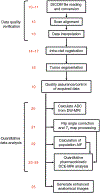
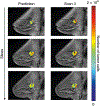
This protocol describes a complete data acquisition, analysis and computational forecasting pipeline for employing quantitative MRI data to predict the response of locally advanced breast cancer to neoadjuvant therapy in a community-based care setting. The methodology has previously been successfully applied to a heterogeneous patient population. The protocol details how to acquire the necessary images followed by registration, segmentation, quantitative perfusion and diffusion analysis, model calibration, and prediction. The data collection portion of the protocol requires ~25 min of scanning, postprocessing requires 2-3 h, and the model calibration and prediction components require ~10 h per patient depending on tumor size. The response of individual breast cancer patients to neoadjuvant therapy is forecast by application of a biophysical, reaction-diffusion mathematical model to these data. Successful application of the protocol results in coregistered MRI data from at least two scan visits that quantifies an individual tumor's size, cellularity and vascular properties. This enables a spatially resolved prediction of how a particular patient's tumor will respond to therapy. Expertise in image acquisition and analysis, as well as the numerical solution of partial differential equations, is required to carry out this protocol.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures





References
Related links
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

