Tissue-Resident-Memory CD8+ T Cells Bridge Innate Immune Responses in Neighboring Epithelial Cells to Control Human Genital Herpes
- PMID: 34552595
- PMCID: PMC8450389
- DOI: 10.3389/fimmu.2021.735643
Tissue-Resident-Memory CD8+ T Cells Bridge Innate Immune Responses in Neighboring Epithelial Cells to Control Human Genital Herpes
Abstract
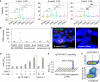
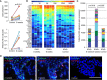
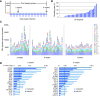
Tissue-resident-memory T cells (TRM) populate the body's barrier surfaces, functioning as frontline responders against reencountered pathogens. Understanding of the mechanisms by which CD8TRM achieve effective immune protection remains incomplete in a naturally recurring human disease. Using laser capture microdissection and transcriptional profiling, we investigate the impact of CD8TRM on the tissue microenvironment in skin biopsies sequentially obtained from a clinical cohort of diverse disease expression during herpes simplex virus 2 (HSV-2) reactivation. Epithelial cells neighboring CD8TRM display elevated and widespread innate and cell-intrinsic antiviral signature expression, largely related to IFNG expression. Detailed evaluation via T-cell receptor reconstruction confirms that CD8TRM recognize viral-infected cells at the specific HSV-2 peptide/HLA level. The hierarchical pattern of core IFN-γ signature expression is well-conserved in normal human skin across various anatomic sites, while elevation of IFI16, TRIM 22, IFITM2, IFITM3, MX1, MX2, STAT1, IRF7, ISG15, IFI44, CXCL10 and CCL5 expression is associated with HSV-2-affected asymptomatic tissue. In primary human cells, IFN-γ pretreatment reduces gene transcription at the immediate-early stage of virus lifecycle, enhances IFI16 restriction of wild-type HSV-2 replication and renders favorable kinetics for host protection. Thus, the adaptive immune response through antigen-specific recognition instructs innate and cell-intrinsic antiviral machinery to control herpes reactivation, a reversal of the canonical thinking of innate activating adaptive immunity in primary infection. Communication from CD8TRM to surrounding epithelial cells to activate broad innate resistance might be critical in restraining various viral diseases.
Keywords: IFI16 restriction factor; cell-intrinsic immunity; human genital herpes; innate antiviral response; tissue microenvironment; tissue-resident-memory T cells (TRM).
Copyright © 2021 Peng, Phasouk, Sodroski, Sun, Hwangbo, Layton, Jin, Klock, Diem, Magaret, Jing, Laing, Li, Huang, Mertens, Johnston, Jerome, Koelle, Wald, Knipe, Corey and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer has declared a shared affiliation with some of the authors CS, MM, DK to the handling Editor at the time of review.
Figures




References
-
- Iwasaki A, Ruslan M. Chapter 8, Innate Responses to Viral Infections. In: Fields Virology. Philadelphia: Lippincott Williams & Wilkins; (2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

