Prognosis Associated With Luminal and Basal Subtypes of Metastatic Prostate Cancer
- PMID: 34554200
- PMCID: PMC8461554
- DOI: 10.1001/jamaoncol.2021.3987
Prognosis Associated With Luminal and Basal Subtypes of Metastatic Prostate Cancer
Abstract
Importance: Luminal and basal subtypes of primary prostate cancer have been shown to be molecularly distinct and clinically important in predicting response to therapy. These subtypes have not been described in metastatic prostate cancer.
Objectives: To identify clinical and molecular correlates of luminal and basal subtypes in metastatic castration-resistant prostate cancer (mCRPC) and investigate differences in survival, particularly after treatment with androgen-signaling inhibitors (ASIs).
Design, setting, and participants: In this cohort study, a retrospective analysis was conducted of 4 cohorts with mCRPC (N = 634) across multiple academic centers. Treatment was at the physicians' discretion. Details of the study cohorts have been published elsewhere between 2016 and 2019. Data were analyzed from March 2018 to February 2021.
Main outcomes and measures: The primary clinical end point was overall survival from the date of tissue biopsy/molecular profiling. Luminal and basal subtypes were also stratified by postbiopsy ASI treatment. The primary molecular analyses included associations with small cell/neuroendocrine prostate cancer (SCNC), molecular pathways, and DNA alterations.
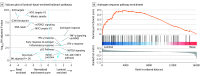
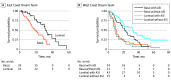
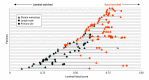
Results: In the 634 patients, 288 (45%) had tumors classified as luminal, and 346 (55%) had tumors classified as basal. However, 53 of 59 (90%) SCNC tumors were basal (P < .001). Similar to primary prostate cancer, luminal tumors exhibited overexpression of AR pathway genes. In basal tumors, a significantly higher rate of RB1 loss (23% basal vs 4% luminal; P < .001), FOXA1 alterations (36% basal vs 27% luminal; P = .03) and MYC alterations (73% basal vs 56% luminal; P < .001) were identified. Patients with basal tumors had worse overall survival compared with those with luminal tumors only in patients treated with an ASI postbiopsy (East Coast Dream Team: hazard ratio [HR], 0.39; 95% CI, 0.20-0.74; P = .004; West Coast Dream Team: HR, 0.57; 95% CI, 0.33-0.97; P = .04). Among patients with luminal tumors, those treated with an ASI had significantly better survival (HR, 0.27; 95% CI, 0.14-0.53; P < .001), whereas patients with basal tumors did not (HR, 0.62; 95% CI, 0.36-1.04, P = .07). The interaction term between subtype and ASI treatment was statistically significant (HR, 0.42; 95% CI, 0.20-0.89; P = .02).
Conclusions and relevance: These findings represent the largest integrated clinical, transcriptomic, and genomic analysis of mCRPC samples to date, and suggest that mCRPC can be classified as luminal and basal tumors. Analogous to primary prostate cancer, these data suggest that the benefit of ASI treatment is more pronounced in luminal tumors and support the use of ASIs in this population. In the basal tumors, a chemotherapeutic approach could be considered in some patients given the similarity to SCNC and the diminished benefit of ASI therapy. Further validation in prospective clinical trials is warranted.
Conflict of interest statement
Figures


Comment in
-
Luminal and Basal Subtypes of Metastatic Prostate Cancer.JAMA Oncol. 2021 Nov 1;7(11):1652-1653. doi: 10.1001/jamaoncol.2021.3970. JAMA Oncol. 2021. PMID: 34554187 No abstract available.
-
Urological Oncology: Prostate Cancer.J Urol. 2022 Apr;207(4):928-930. doi: 10.1097/JU.0000000000002413. Epub 2022 Jan 7. J Urol. 2022. PMID: 34991328 No abstract available.
References
-
- Harris LN, Ismaila N, McShane LM, et al. ; American Society of Clinical Oncology . Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology Clinical Practice guideline. J Clin Oncol. 2016;34(10):1134-1150. doi:10.1200/JCO.2015.65.2289 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

