Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study
- PMID: 34554826
- PMCID: PMC9302186
- DOI: 10.1126/science.abh2315
Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study
Abstract
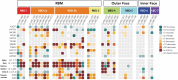
Antibody-based therapeutics and vaccines are essential to combat COVID-19 morbidity and mortality after severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Multiple mutations in SARS-CoV-2 that could impair antibody defenses propagated in human-to-human transmission and spillover or spillback events between humans and animals. To develop prevention and therapeutic strategies, we formed an international consortium to map the epitope landscape on the SARS-CoV-2 spike protein, defining and structurally illustrating seven receptor binding domain (RBD)–directed antibody communities with distinct footprints and competition profiles. Pseudovirion-based neutralization assays reveal spike mutations, individually and clustered together in variants, that affect antibody function among the communities. Key classes of RBD-targeted antibodies maintain neutralization activity against these emerging SARS-CoV-2 variants. These results provide a framework for selecting antibody treatment cocktails and understanding how viral variants might affect antibody therapeutic efficacy.
Figures


References
-
- Piccoli L., Park Y.-J., Tortorici M. A., Czudnochowski N., Walls A. C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L. E., Bowen J. E., Acton O. J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J. C., Mele F., Jovic S., Rodriguez B. F., Gupta S. V., Jin F., Piumatti G., Lo Presti G., Pellanda A. F., Biggiogero M., Tarkowski M., Pizzuto M. S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H. W., Lanzavecchia A., Corti D., Veesler D., Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 183, 1024–1042.e21 (2020). 10.1016/j.cell.2020.09.037 - DOI - PMC - PubMed
-
- Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E. E., Bhattacharya T., Foley B., Hastie K. M., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., McDanal C., Perez L. G., Tang H., Moon-Walker A., Whelan S. P., LaBranche C. C., Saphire E. O., Montefiori D. C.; Sheffield COVID-19 Genomics Group , Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 812–827.e19 (2020). 10.1016/j.cell.2020.06.043 - DOI - PMC - PubMed
-
- Thomson E. C., Rosen L. E., Shepherd J. G., Spreafico R., da Silva Filipe A., Wojcechowskyj J. A., Davis C., Piccoli L., Pascall D. J., Dillen J., Lytras S., Czudnochowski N., Shah R., Meury M., Jesudason N., De Marco A., Li K., Bassi J., O’Toole A., Pinto D., Colquhoun R. M., Culap K., Jackson B., Zatta F., Rambaut A., Jaconi S., Sreenu V. B., Nix J., Zhang I., Jarrett R. F., Glass W. G., Beltramello M., Nomikou K., Pizzuto M., Tong L., Cameroni E., Croll T. I., Johnson N., Di Iulio J., Wickenhagen A., Ceschi A., Harbison A. M., Mair D., Ferrari P., Smollett K., Sallusto F., Carmichael S., Garzoni C., Nichols J., Galli M., Hughes J., Riva A., Ho A., Schiuma M., Semple M. G., Openshaw P. J. M., Fadda E., Baillie J. K., Chodera J. D., Rihn S. J., Lycett S. J., Virgin H. W., Telenti A., Corti D., Robertson D. L., Snell G.; ISARIC4C Investigators; COVID-19 Genomics UK (COG-UK) Consortium , Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187.e20 (2021). 10.1016/j.cell.2021.01.037 - DOI - PMC - PubMed
-
- CDC, Science Brief: Emerging SARS-CoV-2 Variants (2021); www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-b....
-
- Oreshkova N., Molenaar R. J., Vreman S., Harders F., Oude Munnink B. B., Hakze-van der Honing R. W., Gerhards N., Tolsma P., Bouwstra R., Sikkema R. S., Tacken M. G., de Rooij M. M., Weesendorp E., Engelsma M. Y., Bruschke C. J., Smit L. A., Koopmans M., van der Poel W. H., Stegeman A., SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 25, (2020). 10.2807/1560-7917.ES.2020.25.23.2001005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

