The Impact of Real-Time Whole-Genome Sequencing in Controlling Healthcare-Associated SARS-CoV-2 Outbreaks
- PMID: 34555152
- PMCID: PMC8522425
- DOI: 10.1093/infdis/jiab483
The Impact of Real-Time Whole-Genome Sequencing in Controlling Healthcare-Associated SARS-CoV-2 Outbreaks
Abstract
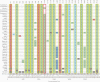
Nosocomial severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections have severely affected bed capacity and patient flow. We utilized whole-genome sequencing (WGS) to identify outbreaks and focus infection control resources and intervention during the United Kingdom's second pandemic wave in late 2020. Phylogenetic analysis of WGS and epidemiological data pinpointed an initial transmission event to an admission ward, with immediate prior community infection linkage documented. High incidence of asymptomatic staff infection with genetically identical viral sequences was also observed, which may have contributed to the propagation of the outbreak. WGS allowed timely nosocomial transmission intervention measures, including admissions ward point-of-care testing and introduction of portable HEPA14 filters. Conversely, WGS excluded nosocomial transmission in 2 instances with temporospatial linkage, conserving time and resources. In summary, WGS significantly enhanced understanding of SARS-CoV-2 clusters in a hospital setting, both identifying high-risk areas and conversely validating existing control measures in other units, maintaining clinical service overall.
Keywords: SARS-CoV-2; cluster; genetic epidemiology; infection control; nosocomial transmission; outbreak; virus; whole-genome sequencing: COVID-19.
© The Author(s) 2021. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures



References
-
- UK Government. Coronavirus in the UK.https://coronavirus.data.gov.uk/. Accessed 21 October 2021.
-
- Berlin DA, Gulick RM, Martinez FJ.. Severe Covid-19. N Engl J Med 2020; 383:2451–60. - PubMed
-
- Gandhi RT, Lynch JB, Del Rio C.. Mild or moderate Covid-19. N Engl J Med 2020; 383:1757–66. - PubMed
-
- Environmental and Modelling Group-Scientific Advisory Group for Emergencies. Transmission of SARS-CoV-2 and mitigating measures - update, 4 June 2020. London, UK: UK Government.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

