SRCP: a comprehensive pipeline for accurate annotation and quantification of circRNAs
- PMID: 34556162
- PMCID: PMC8459468
- DOI: 10.1186/s13059-021-02497-7
SRCP: a comprehensive pipeline for accurate annotation and quantification of circRNAs
Abstract
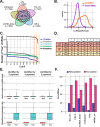
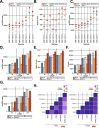
Here we describe a new integrative approach for accurate annotation and quantification of circRNAs named Short Read circRNA Pipeline (SRCP). Our strategy involves two steps: annotation of validated circRNAs followed by a quantification step. We show that SRCP is more sensitive than other individual pipelines and allows for more comprehensive quantification of a larger number of differentially expressed circRNAs. To facilitate the use of SRCP, we generate a comprehensive collection of validated circRNAs in five different organisms, including humans. We then utilize our approach and identify a subset of circRNAs bound to the miRNA-effector protein AGO2 in human brain samples.
Keywords: AGO2; CircRNAs; Circular RNA; Pipeline; RNA metabolism; Splicing.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

