Targeting MYC-enhanced glycolysis for the treatment of small cell lung cancer
- PMID: 34556188
- PMCID: PMC8461854
- DOI: 10.1186/s40170-021-00270-9
Targeting MYC-enhanced glycolysis for the treatment of small cell lung cancer
Abstract
Introduction: The transcription factor MYC is overexpressed in 30% of small cell lung cancer (SCLC) tumors and is known to modulate the balance between two major pathways of metabolism: glycolysis and mitochondrial respiration. This duality of MYC underscores the importance of further investigation into its role in SCLC metabolism and could lead to insights into metabolic targeting approaches.
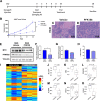
Methods: We investigated differences in metabolic pathways in transcriptional and metabolomics datasets based on cMYC expression in patient and cell line samples. Metabolic pathway utilization was evaluated by flow cytometry and Seahorse extracellular flux methodology. Glycolysis inhibition was evaluated in vitro and in vivo using PFK158, a small molecular inhibitor of PFKFB3.
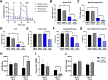
Results: MYC-overexpressing SCLC patient samples and cell lines exhibited increased glycolysis gene expression directly mediated by MYC. Further, MYC-overexpressing cell lines displayed enhanced glycolysis consistent with the Warburg effect, while cell lines with low MYC expression appeared more reliant on oxidative metabolism. Inhibition of glycolysis with PFK158 preferentially attenuated glucose uptake, ATP production, and lactate in MYC-overexpressing cell lines. Treatment with PFK158 in xenografts delayed tumor growth and decreased glycolysis gene expression.
Conclusions: Our study highlights an in-depth characterization of SCLC metabolic programming and presents glycolysis as a targetable mechanism downstream of MYC that could offer therapeutic benefit in a subset of SCLC patients.
Keywords: Glycolysis; MYC; Metabolism; PFK158; Small cell lung cancer.
© 2021. The Author(s).
Conflict of interest statement
L.A.B. serves on advisory committees for AstraZeneca, AbbVie, GenMab, BergenBio, Pharma Mar SA, Sierra Oncology, Merck, Bristol Myers Squibb, Genentech, and Pfizer and has research support from AbbVie, AstraZeneca, GenMab, Sierra Oncology, Tolero Pharmaceuticals. C.M.G. received research funding from AstraZeneca.
Figures



References
-
- N H, AM N, M K, D M, A B, M Y et al. SEER Cancer Statistics Review (CSR) 1975-2016. Bethesda: National Cancer Institute; 2019.
-
- Society AC. Cancer Facts and Figures 2019. Atlanta: American Cancer Society; 2019.
-
- Institute NC . Scientific framework for small cell lung cancer (SCLC) 2014.
Grants and funding
- R01-CA207295/CA/NCI NIH HHS/United States
- U01-CA231844/CA/NCI NIH HHS/United States
- U01 CA235510/CA/NCI NIH HHS/United States
- U01-CA235510/CA/NCI NIH HHS/United States
- S10-OD012304-01/CA/NCI NIH HHS/United States
- T32-CA009666/CA/NCI NIH HHS/United States
- R01 CA207295/CA/NCI NIH HHS/United States
- P50 CA070907/CA/NCI NIH HHS/United States
- U01-CA213274/CA/NCI NIH HHS/United States
- R50-CA243698/CA/NCI NIH HHS/United States
- T32 CA009666/CA/NCI NIH HHS/United States
- U01-CA21373/CA/NCI NIH HHS/United States
- U01 CA213273/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- T32 CA009302/CA/NCI NIH HHS/United States
- P30-CA16672/CA/NCI NIH HHS/United States
- R50 CA243698/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources

