Restriction factor compendium for influenza A virus reveals a mechanism for evasion of autophagy
- PMID: 34556855
- PMCID: PMC9683089
- DOI: 10.1038/s41564-021-00964-2
Restriction factor compendium for influenza A virus reveals a mechanism for evasion of autophagy
Abstract
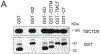
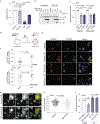
The fate of influenza A virus (IAV) infection in the host cell depends on the balance between cellular defence mechanisms and viral evasion strategies. To illuminate the landscape of IAV cellular restriction, we generated and integrated global genetic loss-of-function screens with transcriptomics and proteomics data. Our multi-omics analysis revealed a subset of both IFN-dependent and independent cellular defence mechanisms that inhibit IAV replication. Amongst these, the autophagy regulator TBC1 domain family member 5 (TBC1D5), which binds Rab7 to enable fusion of autophagosomes and lysosomes, was found to control IAV replication in vitro and in vivo and to promote lysosomal targeting of IAV M2 protein. Notably, IAV M2 was observed to abrogate TBC1D5-Rab7 binding through a physical interaction with TBC1D5 via its cytoplasmic tail. Our results provide evidence for the molecular mechanism utilised by IAV M2 protein to escape lysosomal degradation and traffic to the cell membrane, where it supports IAV budding and growth.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures
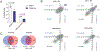



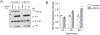






References
-
- Karlas A et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

